You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002355_00450
You are here: Home > Sequence: MGYG000002355_00450
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_F botulinum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_F; Clostridium_F botulinum | |||||||||||
| CAZyme ID | MGYG000002355_00450 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36357; End: 37772 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16800 | Endopep_inhib | 7.13e-41 | 322 | 467 | 1 | 150 | IseA DL-endopeptidase inhibitor. This domain functions as a DL-endopeptidase inhibitor. |
| pfam04122 | CW_binding_2 | 3.15e-26 | 132 | 216 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| 397997 | pfam04122 | 9.51e-26 | 33 | 118 | 1 | 80 | CW_binding_2 Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam04122 | CW_binding_2 | 2.99e-24 | 226 | 313 | 1 | 79 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| NF033435 | S-layer_Clost | 1.67e-23 | 22 | 328 | 382 | 727 | S-layer protein SlpA. In Clostridiodes difficile, the S-layer protein precursor, SlpA, is one member of a large paralogous family of protein that share several cell wall-binding repeats. SlpA is cleaved into a larger and smaller protein. The S-layer protein itself is important to adhesion, and portions of it are highly variable, and then N-terminal and C-terminal are well-conserved. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AAO35771.1 | 2.53e-112 | 1 | 322 | 1 | 321 |
| CDI49287.1 | 3.19e-110 | 1 | 335 | 1 | 328 |
| AKN31997.1 | 9.28e-75 | 25 | 324 | 25 | 323 |
| AKA72454.1 | 7.95e-71 | 17 | 324 | 16 | 323 |
| AWI06417.1 | 1.55e-70 | 17 | 324 | 16 | 323 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J72_A | 1.61e-34 | 32 | 300 | 133 | 438 | ChainA, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630],5J72_B Chain B, Putative N-acetylmuramoyl-L-alanine amidase,autolysin cwp6 [Clostridioides difficile 630] |
| 5J6Q_A | 7.45e-32 | 32 | 306 | 287 | 576 | ChainA, Cell wall binding protein cwp8 [Clostridioides difficile 630] |
| 7ACY_B | 3.32e-25 | 22 | 314 | 38 | 363 | ChainB, S-layer protein [Clostridioides difficile 630],7ACY_D Chain D, S-layer protein [Clostridioides difficile 630],7QGQ_B Chain B, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_D Chain D, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_J Chain J, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_K Chain K, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_L Chain L, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_M Chain M, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_N Chain N, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_T Chain T, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_U Chain U, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_V Chain V, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_W Chain W, Precursor of the S-layer proteins [Clostridioides difficile 630],7QGQ_X Chain X, Precursor of the S-layer proteins [Clostridioides difficile 630] |
| 7ACX_B | 4.52e-25 | 22 | 314 | 38 | 363 | ChainB, S-layer protein [Clostridioides difficile],7ACX_D Chain D, S-layer protein [Clostridioides difficile] |
| 7ACZ_B | 1.93e-21 | 36 | 317 | 45 | 380 | ChainB, SLPH (HMW SLP) [Clostridioides difficile R20291],7ACZ_D Chain D, SLPH (HMW SLP) [Clostridioides difficile R20291] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02114 | 1.71e-52 | 32 | 319 | 29 | 313 | N-acetylmuramoyl-L-alanine amidase LytC OS=Bacillus subtilis (strain 168) OX=224308 GN=lytC PE=1 SV=1 |
| Q02113 | 4.92e-34 | 32 | 350 | 63 | 372 | Amidase enhancer OS=Bacillus subtilis (strain 168) OX=224308 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
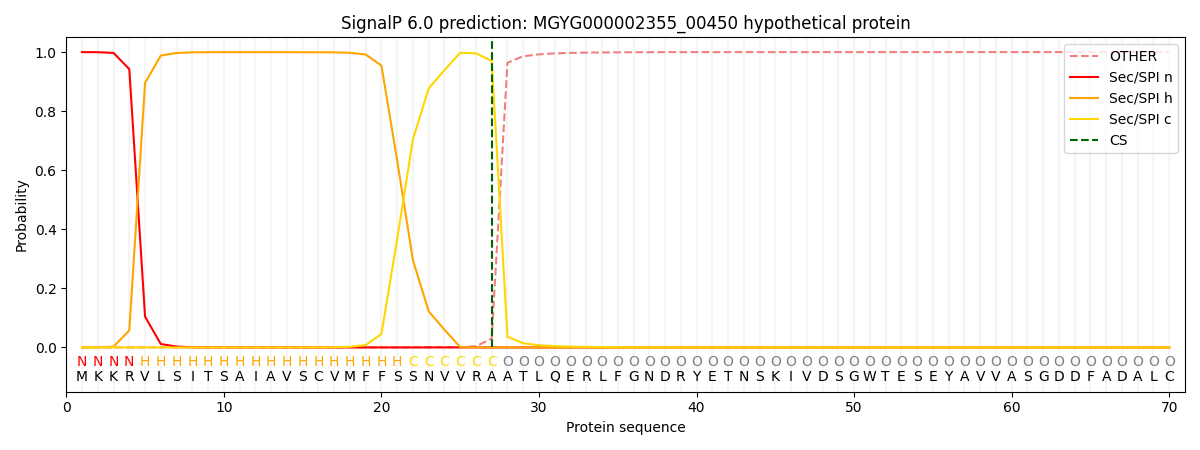
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000489 | 0.998606 | 0.000361 | 0.000201 | 0.000156 | 0.000146 |

