You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002355_01846
You are here: Home > Sequence: MGYG000002355_01846
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_F botulinum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_F; Clostridium_F botulinum | |||||||||||
| CAZyme ID | MGYG000002355_01846 | |||||||||||
| CAZy Family | CBM12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1575553; End: 1577772 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03296 | M6dom_TIGR03296 | 4.59e-46 | 150 | 429 | 1 | 285 | M6 family metalloprotease domain. This model describes a metalloproteinase domain, with a characteristic HExxH motif. Examples of this domain are found in proteins in the family of immune inhibitor A, which cleaves antibacterial peptides, and in other, only distantly related proteases. This model is built to be broader and more inclusive than pfam05547. |
| pfam05547 | Peptidase_M6 | 2.14e-10 | 172 | 419 | 60 | 347 | Immune inhibitor A peptidase M6. The insect pathogenic Gram-positive Bacillus thuringiensis secretes immune inhibitor A, a metallopeptidase, which specifically cleaves host antibacterial proteins. A homolog of immune inhibitor A, PrtV, has been identified in the Gram-negative human pathogen Vibrio cholerae. |
| COG4412 | COG4412 | 1.02e-09 | 173 | 379 | 186 | 430 | Bacillopeptidase F, M6 metalloprotease family [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRI51831.1 | 0.0 | 1 | 627 | 1 | 627 |
| APH17289.1 | 0.0 | 1 | 627 | 1 | 626 |
| ACA55272.1 | 0.0 | 1 | 627 | 1 | 626 |
| APQ76409.1 | 0.0 | 1 | 627 | 1 | 626 |
| AUM99307.1 | 0.0 | 1 | 627 | 1 | 626 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
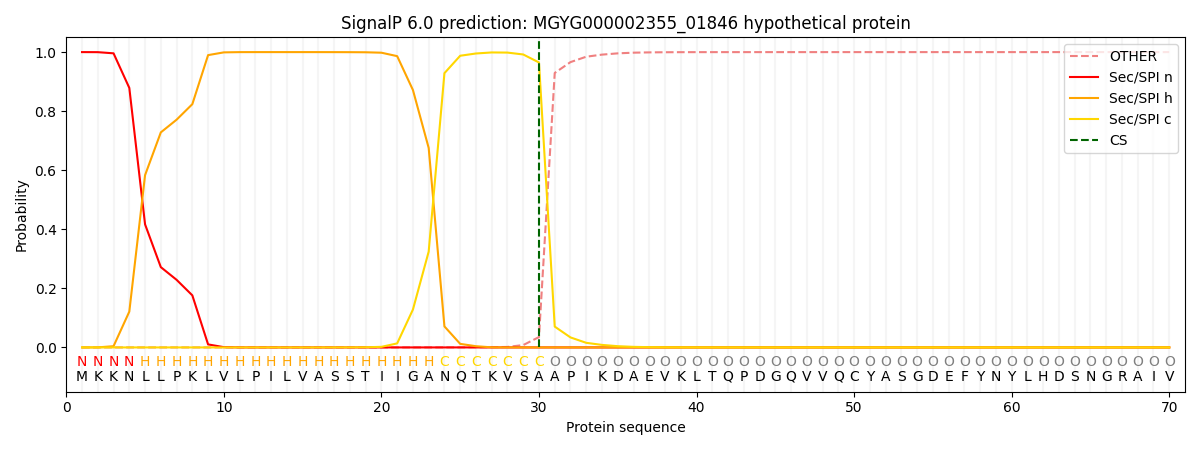
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000287 | 0.998977 | 0.000202 | 0.000202 | 0.000170 | 0.000140 |
