You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002356_01470
You are here: Home > Sequence: MGYG000002356_01470
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
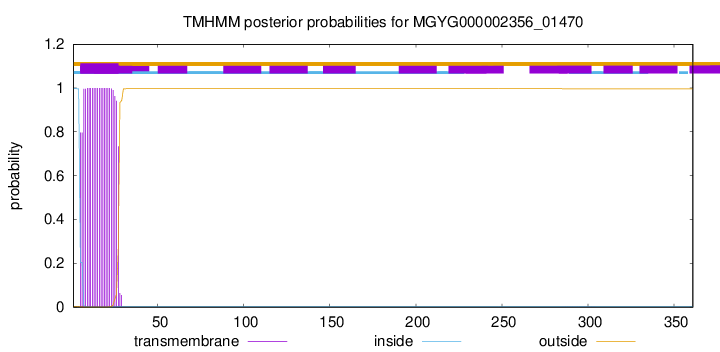
TMHMM annotations
Basic Information help
| Species | Bacillus_A wiedmannii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae_G; Bacillus_A; Bacillus_A wiedmannii | |||||||||||
| CAZyme ID | MGYG000002356_01470 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 293841; End: 294926 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 32 | 352 | 2.7e-106 | 0.9875389408099688 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01270 | Glyco_hydro_8 | 7.63e-04 | 62 | 335 | 28 | 312 | Glycosyl hydrolases family 8. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWH74878.1 | 6.72e-266 | 1 | 361 | 1 | 361 |
| QWI19471.1 | 6.72e-266 | 1 | 361 | 1 | 361 |
| QIW18590.1 | 1.25e-261 | 1 | 361 | 1 | 361 |
| AZQ45502.1 | 1.77e-261 | 1 | 361 | 1 | 361 |
| AYF09141.1 | 7.21e-261 | 1 | 361 | 1 | 361 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6R2M_A | 7.92e-28 | 40 | 319 | 26 | 293 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
| 3REN_A | 2.95e-26 | 55 | 352 | 48 | 344 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 6.63e-59 | 32 | 356 | 45 | 355 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
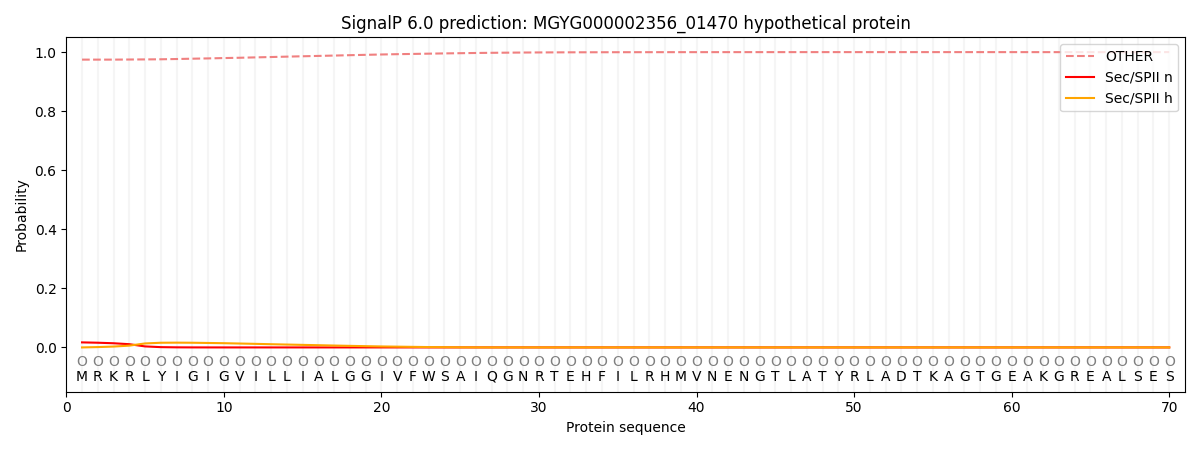
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.974728 | 0.004021 | 0.017098 | 0.000047 | 0.000021 | 0.004088 |