You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002357_00739
You are here: Home > Sequence: MGYG000002357_00739
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus licheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus licheniformis | |||||||||||
| CAZyme ID | MGYG000002357_00739 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | Mannan endo-1,4-beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 742324; End: 743406 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 30 | 348 | 5.2e-92 | 0.9966996699669967 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 2.17e-139 | 30 | 348 | 1 | 311 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 1.23e-95 | 2 | 360 | 6 | 355 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARW45823.1 | 5.38e-276 | 1 | 360 | 1 | 360 |
| AKQ71910.1 | 5.38e-276 | 1 | 360 | 1 | 360 |
| VEH76950.1 | 5.38e-276 | 1 | 360 | 1 | 360 |
| AXF87496.1 | 5.38e-276 | 1 | 360 | 1 | 360 |
| AZN80616.1 | 5.38e-276 | 1 | 360 | 1 | 360 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CBW_A | 5.61e-216 | 17 | 360 | 2 | 345 | ChainA, YdhT protein [Bacillus subtilis],3CBW_B Chain B, YdhT protein [Bacillus subtilis] |
| 2QHA_A | 1.52e-215 | 25 | 360 | 1 | 336 | FromStructure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman [Bacillus subtilis],2QHA_B From Structure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman [Bacillus subtilis] |
| 2WHK_A | 5.76e-214 | 25 | 360 | 1 | 336 | Structureof Bacillus subtilis mannanase man26 [Bacillus subtilis] |
| 7EET_A | 6.86e-116 | 26 | 349 | 9 | 345 | ChainA, Mannanase KMAN [Klebsiella oxytoca] |
| 6Q75_A | 9.27e-30 | 27 | 349 | 21 | 327 | Thestructure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75],6Q75_B The structure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O05512 | 3.15e-217 | 2 | 360 | 3 | 362 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 2.59e-197 | 1 | 360 | 2 | 360 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| P16699 | 3.67e-115 | 6 | 354 | 7 | 360 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| A1A278 | 8.54e-22 | 13 | 322 | 24 | 358 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P49425 | 3.03e-19 | 77 | 292 | 189 | 408 | Mannan endo-1,4-beta-mannosidase OS=Rhodothermus marinus (strain ATCC 43812 / DSM 4252 / R-10) OX=518766 GN=manA PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
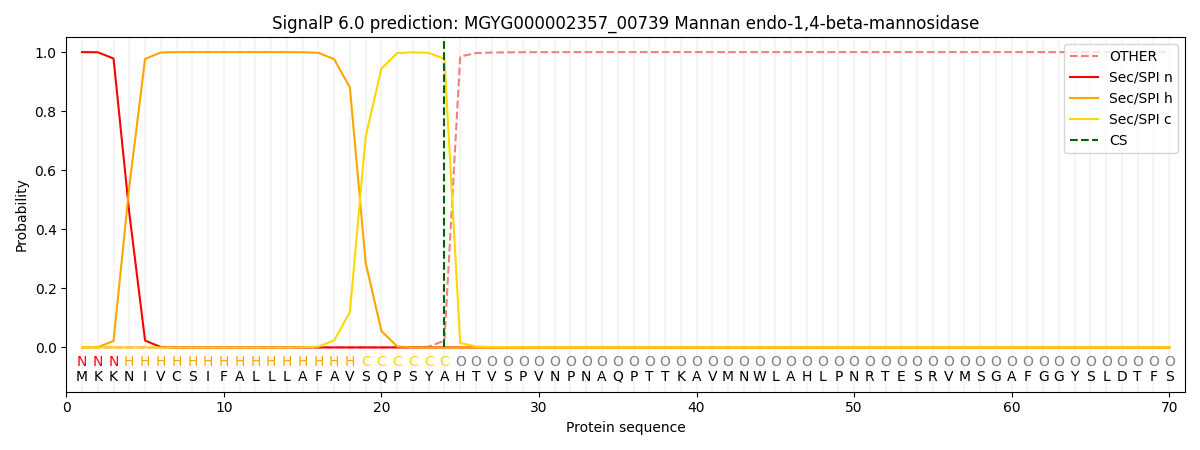
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000269 | 0.999056 | 0.000188 | 0.000167 | 0.000160 | 0.000149 |
