You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002357_03311
You are here: Home > Sequence: MGYG000002357_03311
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
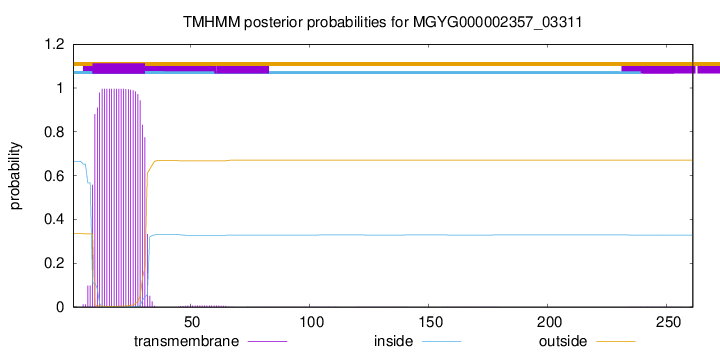
TMHMM annotations
Basic Information help
| Species | Bacillus licheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus licheniformis | |||||||||||
| CAZyme ID | MGYG000002357_03311 | |||||||||||
| CAZy Family | GH12 | |||||||||||
| CAZyme Description | Endoglucanase S | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3175000; End: 3175785 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH12 | 108 | 260 | 3.8e-43 | 0.9935897435897436 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK06215 | PRK06215 | 5.45e-26 | 15 | 259 | 21 | 236 | hypothetical protein; Provisional |
| pfam01670 | Glyco_hydro_12 | 8.61e-25 | 46 | 260 | 3 | 206 | Glycosyl hydrolase family 12. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKI23605.1 | 1.12e-200 | 1 | 261 | 1 | 261 |
| QNT99472.1 | 1.12e-200 | 1 | 261 | 1 | 261 |
| ARC64180.1 | 1.12e-200 | 1 | 261 | 1 | 261 |
| QBR21375.1 | 1.12e-200 | 1 | 261 | 1 | 261 |
| AAU24774.1 | 1.12e-200 | 1 | 261 | 1 | 261 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEM_A | 1.17e-201 | 1 | 261 | 1 | 261 | Nativefamily 12 xyloglucanase from Bacillus licheniformis [Bacillus licheniformis],2JEM_B Native family 12 xyloglucanase from Bacillus licheniformis [Bacillus licheniformis] |
| 2JEN_A | 3.35e-201 | 1 | 261 | 1 | 261 | Family12 xyloglucanase from Bacillus licheniformis in complex with ligand [Bacillus licheniformis] |
| 1OA2_A | 1.11e-26 | 41 | 261 | 10 | 218 | ChainA, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei],1OA2_B Chain B, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei],1OA2_C Chain C, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei],1OA2_D Chain D, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei],1OA2_E Chain E, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei],1OA2_F Chain F, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei] |
| 1OA3_A | 1.11e-26 | 41 | 261 | 10 | 218 | ChainA, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride],1OA3_B Chain B, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride],1OA3_C Chain C, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride],1OA3_D Chain D, ENDO-BETA-1-4-GLUCANASE [Trichoderma citrinoviride] |
| 1OLQ_A | 1.11e-26 | 41 | 261 | 10 | 218 | ChainA, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei],1OLQ_B Chain B, ENDO-BETA-1,4-GLUCANASE [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16630 | 2.91e-122 | 15 | 261 | 18 | 264 | Endoglucanase S OS=Pectobacterium parmentieri OX=1905730 GN=celS PE=1 SV=1 |
| Q5BG78 | 2.20e-18 | 48 | 260 | 37 | 238 | Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
| G4N5V2 | 8.84e-18 | 40 | 261 | 44 | 263 | Endoglucanase cel12B OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=cel12B PE=1 SV=1 |
| Q12679 | 1.56e-17 | 25 | 261 | 7 | 239 | Endoglucanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=cekA PE=2 SV=2 |
| W2PEP3 | 2.09e-15 | 20 | 261 | 4 | 241 | Xyloglucan-specific endo-beta-1,4-glucanase 1 OS=Phytophthora parasitica (strain INRA-310) OX=761204 GN=XEG1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
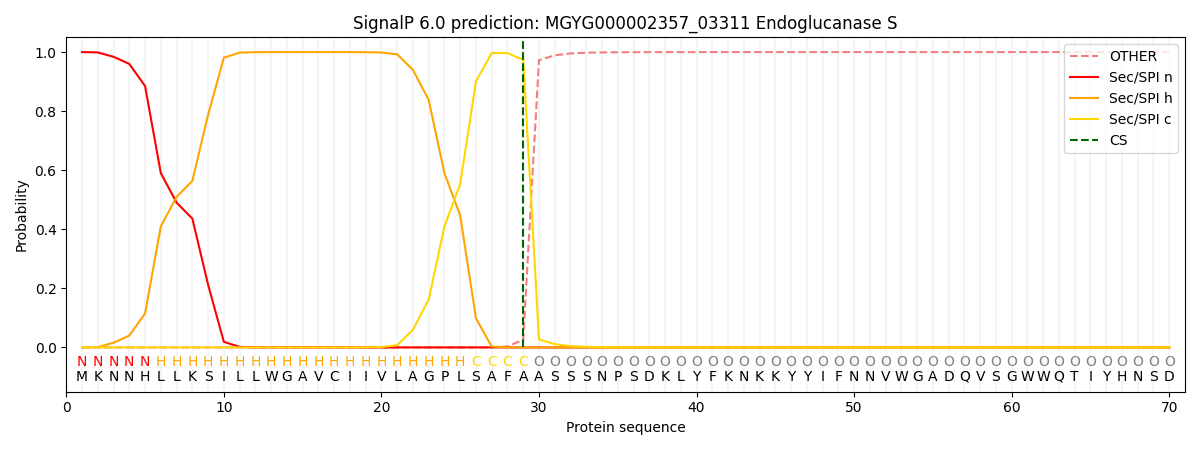
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000408 | 0.998855 | 0.000213 | 0.000184 | 0.000163 | 0.000159 |