You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002362_00877
You are here: Home > Sequence: MGYG000002362_00877
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
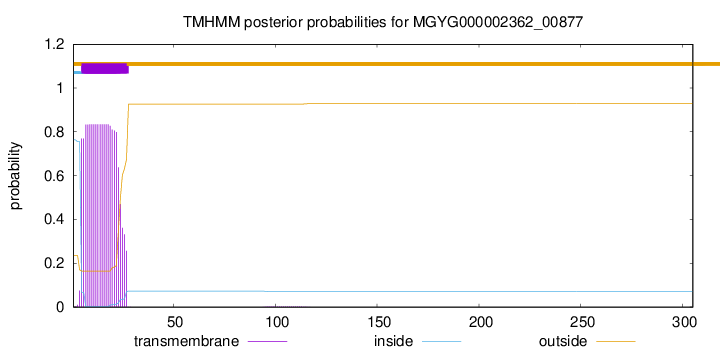
TMHMM annotations
Basic Information help
| Species | Fusobacterium_C necrophorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium_C; Fusobacterium_C necrophorum | |||||||||||
| CAZyme ID | MGYG000002362_00877 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 103228; End: 104145 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2342 | COG2342 | 7.22e-58 | 133 | 305 | 2 | 157 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
| TIGR01370 | TIGR01370 | 5.59e-29 | 185 | 304 | 59 | 177 | extracellular protein. Original assignment of this protein family as cysteinyl-tRNA synthetase is controversial, supported by but challenged by and by subsequent discovery of the actual mechanism for synthesizing Cys-tRNA in species where a direct Cys--tRNA ligase was not found. Lingering legacy annotations of members of this family probably should be removed. Evidence against the role includes a signal peptide. This family as been renamed "extracellular protein" to facilitate correction. Members of this family occur in Deinococcus radiodurans (bacterial) and Methanococcus jannaschii (archaeal). A number of homologous but more distantly related proteins are annotated as alpha-1,4 polygalactosaminidases. The function remains unknown. [Unknown function, General] |
| pfam03537 | Glyco_hydro_114 | 1.92e-13 | 206 | 299 | 28 | 112 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG2342 | COG2342 | 0.010 | 31 | 115 | 176 | 252 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYV94831.1 | 1.38e-223 | 1 | 305 | 1 | 305 |
| AVQ21050.1 | 2.78e-223 | 1 | 305 | 1 | 305 |
| AYV92755.1 | 2.78e-223 | 1 | 305 | 1 | 305 |
| AZW09275.1 | 1.65e-136 | 1 | 305 | 1 | 305 |
| AYZ72730.1 | 1.65e-136 | 1 | 305 | 1 | 305 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2AAM_A | 3.41e-23 | 186 | 305 | 35 | 153 | Crystalstructure of a putative glycosidase (tm1410) from thermotoga maritima at 2.20 A resolution [Thermotoga maritima],2AAM_B Crystal structure of a putative glycosidase (tm1410) from thermotoga maritima at 2.20 A resolution [Thermotoga maritima],2AAM_C Crystal structure of a putative glycosidase (tm1410) from thermotoga maritima at 2.20 A resolution [Thermotoga maritima],2AAM_D Crystal structure of a putative glycosidase (tm1410) from thermotoga maritima at 2.20 A resolution [Thermotoga maritima],2AAM_E Crystal structure of a putative glycosidase (tm1410) from thermotoga maritima at 2.20 A resolution [Thermotoga maritima],2AAM_F Crystal structure of a putative glycosidase (tm1410) from thermotoga maritima at 2.20 A resolution [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q58872 | 3.72e-26 | 156 | 305 | 55 | 201 | Uncharacterized protein MJ1477 OS=Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) OX=243232 GN=MJ1477 PE=4 SV=1 |
| Q9X1D0 | 2.30e-22 | 186 | 305 | 49 | 167 | Uncharacterized protein TM_1410 OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=TM_1410 PE=1 SV=1 |
| Q9RWG3 | 7.00e-15 | 216 | 299 | 94 | 177 | Uncharacterized protein DR_0705 OS=Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / LMG 4051 / NBRC 15346 / NCIMB 9279 / R1 / VKM B-1422) OX=243230 GN=DR_0705 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
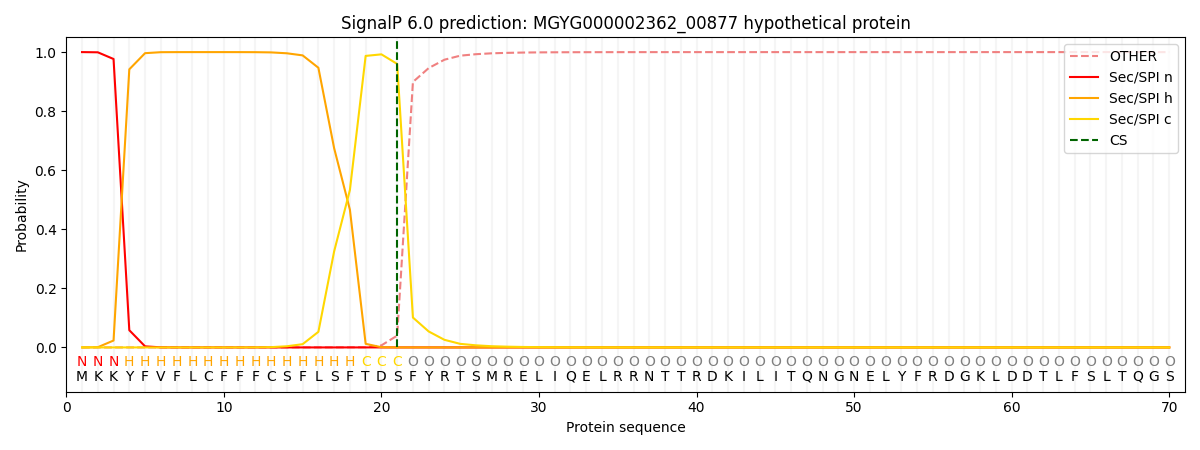
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000373 | 0.998818 | 0.000293 | 0.000166 | 0.000168 | 0.000163 |