You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002368_03313
You are here: Home > Sequence: MGYG000002368_03313
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
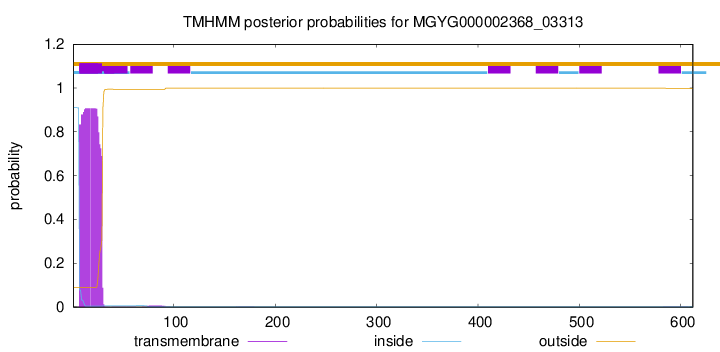
TMHMM annotations
Basic Information help
| Species | Vibrio metoecus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Vibrionaceae; Vibrio; Vibrio metoecus | |||||||||||
| CAZyme ID | MGYG000002368_03313 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | Hemagglutinin/proteinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9152; End: 10990 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3227 | LasB | 0.0 | 1 | 502 | 1 | 507 | Zn-dependent metalloprotease [Posttranslational modification, protein turnover, chaperones]. |
| cd09597 | M4_TLP | 1.13e-94 | 241 | 501 | 1 | 278 | Peptidase M4 family including thermolysin, protealysin, aureolysin, and neutral protease. This peptidase M4 family includes several endopeptidases such as thermolysin (EC 3.4.24.27), aureolysin (the extracellular metalloproteinase from Staphylococcus aureus), neutral protease from Bacillus cereus, protealysin, and bacillolysin (EC 3.4.24.28). Typically, the M4 peptidases consist of a presequence (signal sequence), a propeptide sequence, and a peptidase unit. The presequence is cleaved off during export while the propeptide has inhibitory and chaperone functions and facilitates folding. The propeptide remains attached until the peptidase is secreted and can be safely activated. All peptidases in this family bind a single catalytic zinc ion which is tetrahedrally co-ordinated by three amino acid ligands and a water molecule that forms the nucleophile on activation during catalysis. The active site is found between two sub-domains; the N-terminal domain contains the HEXXH zinc-binding motif while the helical C-terminal domain, which is unique for the family, carries the third zinc ligand. These peptidases are secreted eubacterial endopeptidases from Gram-positive or Gram-negative sources that degrade extracellular proteins and peptides for bacterial nutrition. They are selectively inhibited by Steptomyces metalloproteinase inhibitor (SMPI) as well as by phosphoramidon from Streptomyces tanashiensis. A large number of these enzymes are implicated as key factors in the pathogenesis of various diseases, including gastritis, peptic ulcer, gastric carcinoma, cholera and several types of bacterial infections, and are therefore important drug targets. Some enzymes of the family can function at extremes of temperatures, while some function in organic solvents, thus rendering them novel targets for biotechnological applications. Thermolysin is widely used as a nonspecific protease to obtain fragments for peptide sequencing. It has also been used in production of the artificial sweetener aspartame. |
| pfam01447 | Peptidase_M4 | 5.92e-45 | 211 | 354 | 1 | 147 | Thermolysin metallopeptidase, catalytic domain. |
| pfam02868 | Peptidase_M4_C | 1.58e-42 | 357 | 501 | 1 | 167 | Thermolysin metallopeptidase, alpha-helical domain. |
| cd02699 | M4_M36 | 6.80e-22 | 281 | 500 | 41 | 313 | Peptidase M4 family (includes thermolysin, aureolysin, neutral protease and bacillolysin) and Peptidase M36 family (also known as fungalysin). This family includes the peptidases M4 as well as M36, both belonging to the Gluzincin family. The M4 peptidase family includes numerous zinc-dependent metallopeptidases that hydrolyze peptide bonds, such as thermolysin (EC 3.4.24.27), pseudolysin (the extracellullar elastase of Pseudomonas aeruginosa), aureolysin (the extracellular metalloproteinase from Staphylococcus aureus), neutral protease from Bacillus cereus, as well as bacillolysin (EC 3.4.24.28). The M36 family also known as fungalysin (elastinolytic metalloproteinase) family, includes endopeptidases from pathogenic fungi. Both M4 and M36 families have similar folds and contain the Zn-binding site and the active site HEXXH motif. The eukaryotic M36 and bacterial M4 families of metalloproteases also share a conserved domain in their propeptides called FTP (fungalysin/thermolysin propeptide). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAE6907070.1 | 3.20e-148 | 9 | 612 | 3 | 610 |
| QUM84093.1 | 2.10e-142 | 65 | 612 | 59 | 610 |
| QUM88395.1 | 1.17e-141 | 65 | 612 | 59 | 610 |
| QUM79862.1 | 3.30e-141 | 65 | 612 | 59 | 610 |
| AZQ09362.1 | 1.10e-140 | 24 | 504 | 24 | 497 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7ECC_A | 3.22e-137 | 201 | 517 | 1 | 318 | ChainA, M4 family peptidase [Pseudoalteromonas lipolytica SCSIO 04301] |
| 3NQY_B | 3.13e-132 | 201 | 511 | 1 | 312 | Crystalstructure of the autoprocessed complex of Vibriolysin MCP-02 with a single point mutation E346A [Pseudoalteromonas sp. SM9913] |
| 3NQZ_B | 3.13e-132 | 201 | 511 | 1 | 312 | Crystalstructure of the autoprocessed Vibriolysin MCP-02 with E369A mutation [Pseudoalteromonas sp. SM9913] |
| 3NQX_A | 6.48e-132 | 201 | 504 | 1 | 299 | Crystalstructure of vibriolysin MCP-02 mature enzyme, a zinc metalloprotease from M4 family [Pseudoalteromonas sp. SM9913] |
| 7OC7_A | 1.44e-126 | 201 | 501 | 1 | 295 | ChainA, Neutral metalloproteinase [Pseudomonas aeruginosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P43147 | 0.0 | 1 | 612 | 1 | 611 | Virulence metalloprotease OS=Vibrio anguillarum OX=55601 GN=empA PE=1 SV=1 |
| P24153 | 0.0 | 1 | 612 | 1 | 609 | Hemagglutinin/proteinase OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=hap PE=1 SV=1 |
| Q00971 | 1.66e-318 | 1 | 612 | 1 | 609 | Neutral protease OS=Vibrio proteolyticus OX=671 GN=nprV PE=1 SV=1 |
| Q48259 | 3.64e-177 | 356 | 612 | 1 | 256 | Zinc metalloprotease (Fragment) OS=Helicobacter pylori OX=210 GN=hap PE=3 SV=1 |
| Q02RJ6 | 5.25e-155 | 52 | 501 | 56 | 492 | Elastase OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=lasB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
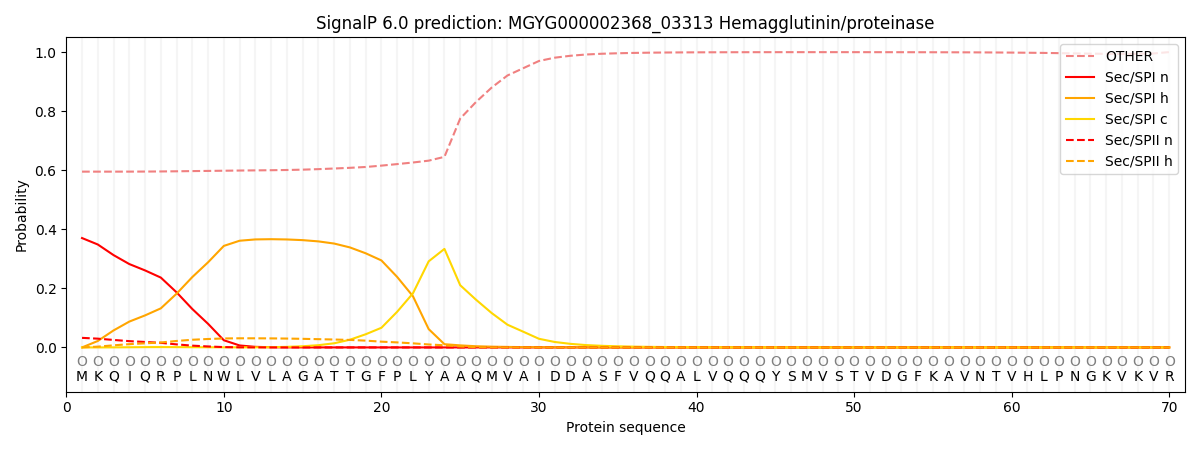
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.614089 | 0.346581 | 0.033878 | 0.001541 | 0.000747 | 0.003154 |