You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002372_00222
You are here: Home > Sequence: MGYG000002372_00222
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_P perfringens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_P; Clostridium_P perfringens | |||||||||||
| CAZyme ID | MGYG000002372_00222 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | Hyaluronoglucosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 252335; End: 257221 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 187 | 491 | 4e-104 | 0.976271186440678 |
| CBM32 | 816 | 941 | 8.6e-18 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 1.17e-120 | 187 | 500 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| cd00057 | FA58C | 5.70e-27 | 800 | 934 | 1 | 134 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 5.12e-17 | 813 | 941 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02838 | Glyco_hydro_20b | 4.94e-16 | 39 | 180 | 2 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| cd14254 | Dockerin_II | 4.14e-10 | 1578 | 1627 | 1 | 54 | Type II dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type II dockerins, which are responsible for mediating attachment of the cellulosome complex to the bacterial cell wall. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOY52617.1 | 0.0 | 1 | 1628 | 1 | 1628 |
| QQA12181.1 | 0.0 | 1 | 1628 | 1 | 1628 |
| ASY50288.1 | 0.0 | 1 | 1628 | 1 | 1628 |
| QUD73312.1 | 0.0 | 1 | 1628 | 1 | 1628 |
| SQG37474.1 | 0.0 | 1 | 1628 | 1 | 1628 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV4_A | 0.0 | 30 | 660 | 23 | 653 | Structureof CpGH84A [Clostridium perfringens ATCC 13124],6PV4_B Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_C Structure of CpGH84A [Clostridium perfringens ATCC 13124],6PV4_D Structure of CpGH84A [Clostridium perfringens ATCC 13124] |
| 6PWI_A | 1.23e-140 | 39 | 649 | 34 | 622 | Structureof CpGH84D [Clostridium perfringens ATCC 13124],6PWI_B Structure of CpGH84D [Clostridium perfringens ATCC 13124] |
| 2W1Q_A | 2.85e-108 | 807 | 975 | 24 | 192 | Uniqueligand binding specificity for a family 32 Carbohydrate- Binding Module from the Mu toxin produced by Clostridium perfringens [Clostridium perfringens],2W1Q_B Unique ligand binding specificity for a family 32 Carbohydrate- Binding Module from the Mu toxin produced by Clostridium perfringens [Clostridium perfringens],2W1U_A A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc [Clostridium perfringens],2W1U_B A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc [Clostridium perfringens],2W1U_C A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc [Clostridium perfringens],2W1U_D A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,3) galNAc [Clostridium perfringens],2WDB_A A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose [Clostridium perfringens],2WDB_B A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose [Clostridium perfringens],2WDB_C A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose [Clostridium perfringens],2WDB_D A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose [Clostridium perfringens] |
| 2W1S_A | 1.91e-107 | 807 | 975 | 24 | 192 | Uniqueligand binding specificity of a family 32 Carbohydrate-Binding Module from the Mu toxin produced by Clostridium perfringens [Clostridium perfringens],2W1S_B Unique ligand binding specificity of a family 32 Carbohydrate-Binding Module from the Mu toxin produced by Clostridium perfringens [Clostridium perfringens] |
| 4TXW_A | 1.23e-106 | 1063 | 1229 | 7 | 173 | Crystalstructure of CBM32-4 from the Clostridium perfringens NagH [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 0.0 | 1 | 1628 | 1 | 1628 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| Q89ZI2 | 9.04e-55 | 82 | 592 | 53 | 535 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| Q8XL08 | 2.02e-51 | 43 | 616 | 48 | 601 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q0TR53 | 8.23e-51 | 43 | 824 | 48 | 795 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| O60502 | 8.85e-22 | 188 | 461 | 63 | 331 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
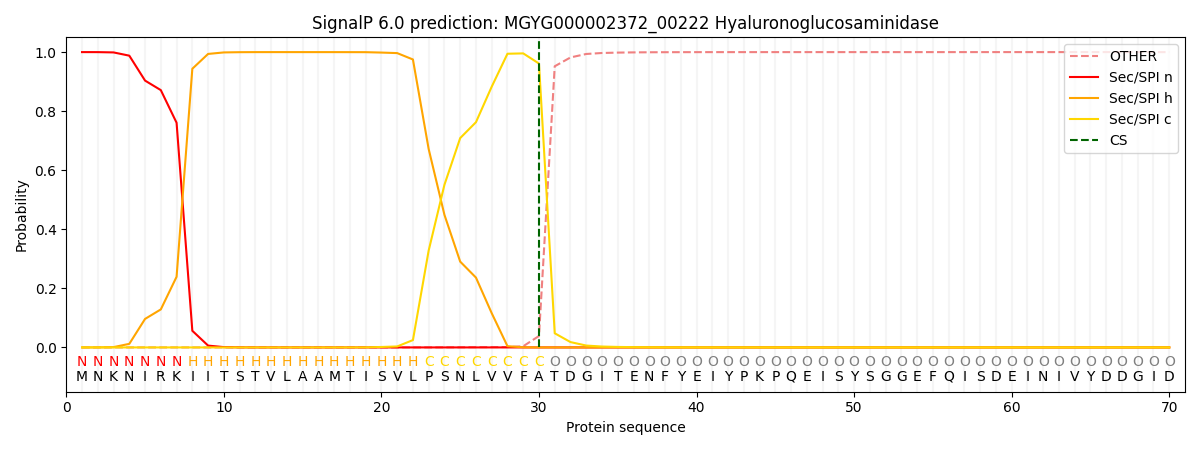
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000323 | 0.998915 | 0.000195 | 0.000207 | 0.000177 | 0.000159 |

