You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002373_00977
You are here: Home > Sequence: MGYG000002373_00977
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
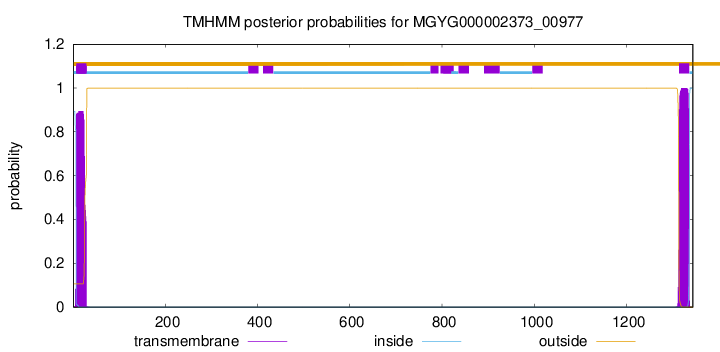
TMHMM annotations
Basic Information help
| Species | Clostridium septicum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium septicum | |||||||||||
| CAZyme ID | MGYG000002373_00977 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 96846; End: 100880 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 36 | 596 | 3.2e-98 | 0.9918533604887984 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 1.63e-05 | 276 | 410 | 5 | 117 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 5.52e-04 | 280 | 381 | 55 | 140 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| COG1196 | Smc | 0.004 | 1078 | 1279 | 218 | 421 | Chromosome segregation ATPase [Cell cycle control, cell division, chromosome partitioning]. |
| cd14251 | PL-6 | 0.005 | 420 | 520 | 261 | 369 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| TIGR02168 | SMC_prok_B | 0.005 | 1016 | 1282 | 685 | 938 | chromosome segregation protein SMC, common bacterial type. SMC (structural maintenance of chromosomes) proteins bind DNA and act in organizing and segregating chromosomes for partition. SMC proteins are found in bacteria, archaea, and eukaryotes. This family represents the SMC protein of most bacteria. The smc gene is often associated with scpB (TIGR00281) and scpA genes, where scp stands for segregation and condensation protein. SMC was shown (in Caulobacter crescentus) to be induced early in S phase but present and bound to DNA throughout the cell cycle. [Cellular processes, Cell division, DNA metabolism, Chromosome-associated proteins] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYE35145.1 | 0.0 | 1 | 1340 | 1 | 1340 |
| AYE35146.1 | 3.20e-289 | 862 | 1344 | 973 | 1455 |
| AZG68471.1 | 1.61e-143 | 20 | 503 | 96 | 566 |
| QNK55089.1 | 2.10e-63 | 33 | 600 | 251 | 739 |
| QNK55088.1 | 2.72e-63 | 32 | 662 | 54 | 603 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 1.55e-28 | 30 | 583 | 3 | 563 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 3.96e-21 | 26 | 531 | 8 | 517 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 1.09e-18 | 30 | 517 | 8 | 514 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 3.67e-18 | 26 | 436 | 235 | 619 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 2.29e-16 | 26 | 436 | 235 | 619 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 2.92e-13 | 1081 | 1340 | 1667 | 1932 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| Q9L7Q2 | 1.89e-10 | 1006 | 1241 | 414 | 660 | Zinc metalloprotease ZmpB OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=zmpB PE=3 SV=2 |
| P26831 | 4.02e-06 | 1042 | 1204 | 1388 | 1561 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
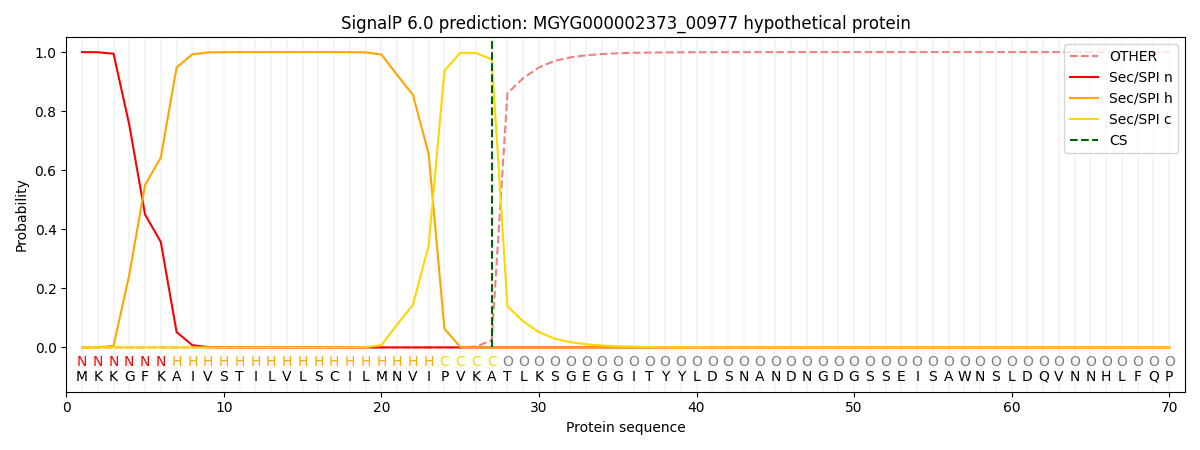
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000352 | 0.998868 | 0.000244 | 0.000181 | 0.000157 | 0.000150 |