You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002373_01185
You are here: Home > Sequence: MGYG000002373_01185
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
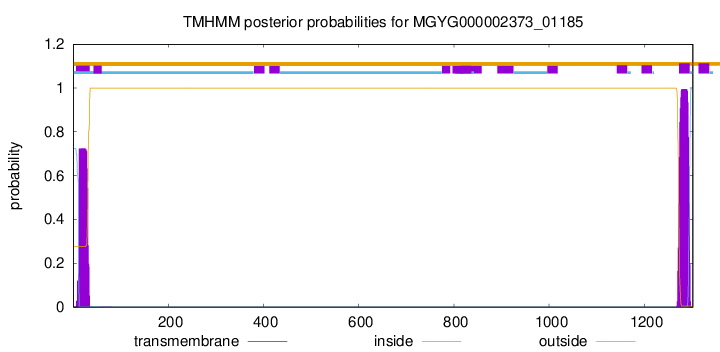
TMHMM annotations
Basic Information help
| Species | Clostridium septicum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium septicum | |||||||||||
| CAZyme ID | MGYG000002373_01185 | |||||||||||
| CAZy Family | GH84 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18953; End: 22861 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH84 | 193 | 491 | 1.4e-108 | 0.9932203389830508 |
| CBM32 | 663 | 767 | 4.6e-16 | 0.8064516129032258 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07555 | NAGidase | 2.89e-140 | 193 | 490 | 1 | 293 | beta-N-acetylglucosaminidase. This family has previously been described as a hyaluronidase. However, more recently it has been shown that this family has beta-N-acetylglucosaminidase activity. |
| pfam02838 | Glyco_hydro_20b | 1.27e-20 | 49 | 186 | 4 | 123 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| pfam00754 | F5_F8_type_C | 3.36e-11 | 663 | 766 | 9 | 118 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 2.14e-09 | 900 | 1019 | 3 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.05e-05 | 676 | 757 | 32 | 125 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYE33864.1 | 0.0 | 1 | 1302 | 1 | 1302 |
| QAS62007.1 | 0.0 | 1 | 1302 | 1 | 1302 |
| AMN32340.1 | 0.0 | 1 | 1301 | 1 | 1296 |
| ABG84510.1 | 0.0 | 1 | 1301 | 1 | 1296 |
| AQW26168.1 | 0.0 | 1 | 1301 | 1 | 1296 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PV5_A | 5.37e-283 | 37 | 625 | 30 | 621 | Structureof CpGH84B [Clostridium perfringens ATCC 13124] |
| 2V5D_A | 1.98e-173 | 51 | 770 | 18 | 713 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
| 2CBI_A | 5.57e-170 | 51 | 623 | 18 | 586 | Structureof the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase [Clostridium perfringens],2CBI_B Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase [Clostridium perfringens],2CBJ_A Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc [Clostridium perfringens],2CBJ_B Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase in complex with PUGNAc [Clostridium perfringens],2V5C_A Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure [Clostridium perfringens],2V5C_B Family 84 glycoside hydrolase from Clostridium perfringens, 2.1 Angstrom structure [Clostridium perfringens],2VUR_A Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death [Clostridium perfringens],2VUR_B Chemical dissection of the link between Streptozotocin, O-GlcNAc and pancreatic cell death [Clostridium perfringens],2X0Y_A Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds [Clostridium perfringens],2X0Y_B Screening-based discovery of drug-like O-GlcNAcase inhibitor scaffolds [Clostridium perfringens] |
| 5OXD_A | 6.85e-170 | 51 | 623 | 20 | 588 | Complexof a C. perfringens O-GlcNAcase with a fragment hit [Clostridium perfringens] |
| 2J62_A | 7.81e-170 | 51 | 623 | 18 | 586 | Structureof a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2J62_B Structure of a bacterial O-glcnacase in complex with glcnacstatin [Clostridium perfringens],2WB5_A GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens],2WB5_B GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 1.10e-171 | 51 | 875 | 48 | 843 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 5.93e-171 | 51 | 875 | 48 | 843 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q89ZI2 | 8.59e-103 | 44 | 783 | 21 | 736 | O-GlcNAcase BT_4395 OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=BT_4395 PE=1 SV=1 |
| P26831 | 1.07e-62 | 48 | 1022 | 40 | 1059 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| O60502 | 7.43e-37 | 192 | 459 | 61 | 331 | Protein O-GlcNAcase OS=Homo sapiens OX=9606 GN=OGA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
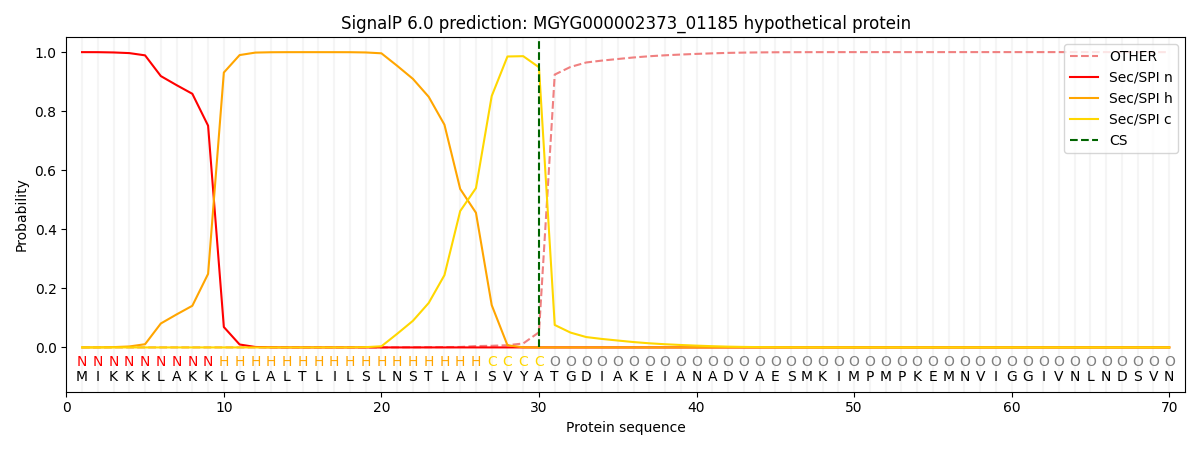
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000723 | 0.998479 | 0.000272 | 0.000179 | 0.000160 | 0.000164 |