You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002378_01638
You are here: Home > Sequence: MGYG000002378_01638
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
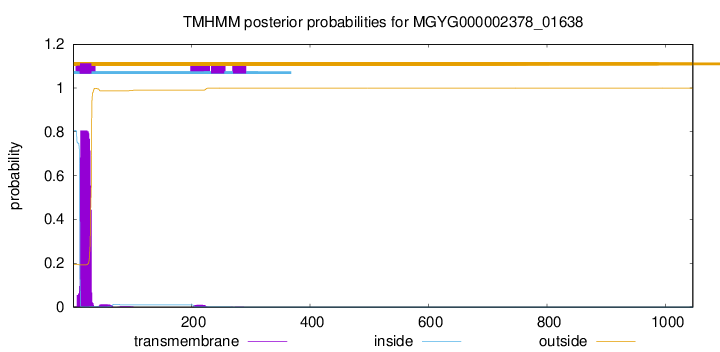
TMHMM annotations
Basic Information help
| Species | Akkermansia sp001580195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae; Akkermansia; Akkermansia sp001580195 | |||||||||||
| CAZyme ID | MGYG000002378_01638 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 132128; End: 135268 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 355 | 726 | 8.9e-79 | 0.9590643274853801 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 4.54e-79 | 362 | 726 | 14 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| cd14948 | BACON | 8.92e-18 | 743 | 819 | 9 | 83 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
| pfam13859 | BNR_3 | 1.91e-16 | 364 | 621 | 1 | 212 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam13088 | BNR_2 | 1.95e-16 | 398 | 709 | 23 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| pfam14873 | BNR_assoc_N | 3.96e-15 | 191 | 325 | 8 | 149 | N-terminal domain of BNR-repeat neuraminidase. This domain is usually found at the N-terminus of the BNR-repeat neuraminidase protein family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWP54611.1 | 0.0 | 2 | 1045 | 1 | 1044 |
| QWP27708.1 | 0.0 | 2 | 1045 | 1 | 1044 |
| QWP61791.1 | 0.0 | 2 | 1045 | 1 | 1044 |
| QWP69127.1 | 0.0 | 2 | 1045 | 1 | 1044 |
| QWP03917.1 | 0.0 | 2 | 1045 | 1 | 1044 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6MYV_A | 1.16e-68 | 295 | 712 | 121 | 504 | Sialidase26co-crystallized with DANA-Gc [bacterium],6MYV_B Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_C Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_D Sialidase26 co-crystallized with DANA-Gc [bacterium] |
| 6MRV_A | 2.19e-68 | 295 | 712 | 142 | 525 | Sialidase26co-crystallized with DANA [bacterium],6MRV_B Sialidase26 co-crystallized with DANA [bacterium],6MRX_A Sialidase26 apo [bacterium],6MRX_B Sialidase26 apo [bacterium],6MRX_C Sialidase26 apo [bacterium],6MRX_D Sialidase26 apo [bacterium] |
| 6MNJ_A | 1.32e-67 | 288 | 712 | 135 | 521 | Hadzamicrobial sialidase Hz136 [Alistipes],6MNJ_B Hadza microbial sialidase Hz136 [Alistipes] |
| 4FJ6_A | 3.41e-64 | 295 | 712 | 122 | 504 | Crystalstructure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_B Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_C Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_D Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503] |
| 4Q6K_A | 3.36e-61 | 295 | 712 | 124 | 507 | Crystalstructure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_B Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_C Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_D Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P31206 | 2.03e-65 | 195 | 712 | 34 | 526 | Sialidase OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=nanH PE=3 SV=2 |
| Q02834 | 3.87e-30 | 331 | 712 | 31 | 386 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P15698 | 6.08e-21 | 362 | 717 | 54 | 384 | Sialidase OS=Paeniclostridium sordellii OX=1505 PE=1 SV=1 |
| P10481 | 1.70e-17 | 359 | 732 | 33 | 374 | Sialidase OS=Clostridium perfringens OX=1502 GN=nanH PE=1 SV=1 |
| Q5RAF4 | 4.06e-13 | 362 | 729 | 77 | 395 | Sialidase-1 OS=Pongo abelii OX=9601 GN=NEU1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
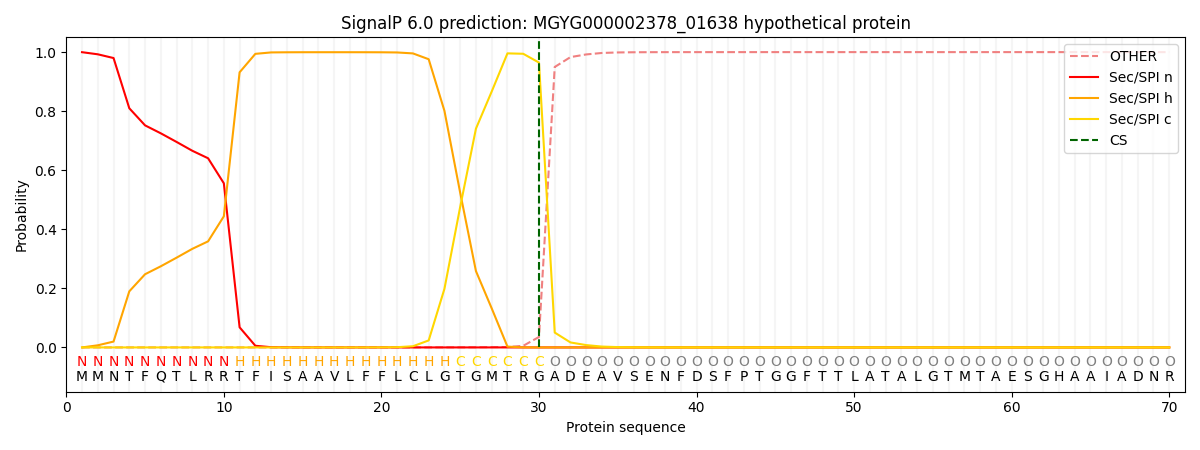
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000575 | 0.998068 | 0.000731 | 0.000218 | 0.000188 | 0.000170 |