You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002381_03574
You are here: Home > Sequence: MGYG000002381_03574
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas bentonitica_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas bentonitica_A | |||||||||||
| CAZyme ID | MGYG000002381_03574 | |||||||||||
| CAZy Family | PL6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 163431; End: 165656 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL6 | 35 | 405 | 1.4e-134 | 0.989247311827957 |
| PL6 | 479 | 679 | 1.4e-41 | 0.5850144092219021 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14251 | PL-6 | 2.59e-172 | 50 | 406 | 14 | 369 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam14592 | Chondroitinas_B | 9.42e-125 | 52 | 447 | 16 | 420 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOX62678.1 | 0.0 | 13 | 741 | 4 | 732 |
| QIO86637.1 | 0.0 | 23 | 741 | 5 | 723 |
| QHB73993.1 | 0.0 | 13 | 741 | 2 | 730 |
| AHY60627.1 | 0.0 | 23 | 741 | 5 | 723 |
| ASR42253.1 | 0.0 | 21 | 741 | 6 | 726 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GKD_A | 1.03e-207 | 33 | 716 | 1 | 690 | Structureof PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_B Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_C Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_D Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis] |
| 5GKQ_A | 8.23e-207 | 33 | 716 | 1 | 690 | Structureof PL6 family alginate lyase AlyGC mutant-R241A [Paraglaciecola chathamensis S18K6],5GKQ_B Structure of PL6 family alginate lyase AlyGC mutant-R241A [Paraglaciecola chathamensis S18K6] |
| 7O7T_A | 3.13e-164 | 36 | 716 | 3 | 690 | ChainA, Poly(Beta-D-mannuronate) lyase [Pseudoalteromonas atlantica T6c] |
| 7O77_A | 3.13e-164 | 36 | 716 | 3 | 690 | ChainA, Poly(Beta-D-mannuronate) lyase [Pseudoalteromonas atlantica T6c] |
| 7DMK_A | 1.16e-116 | 52 | 715 | 29 | 679 | ChainA, BcAlyPL6 [Bacteroides clarus],7DMK_B Chain B, BcAlyPL6 [Bacteroides clarus],7DMK_C Chain C, BcAlyPL6 [Bacteroides clarus],7DMK_D Chain D, BcAlyPL6 [Bacteroides clarus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q06365 | 3.08e-74 | 52 | 379 | 2 | 323 | Alginate lyase OS=Pseudomonas sp. (strain OS-ALG-9) OX=86038 GN=aly PE=3 SV=1 |
| Q46079 | 4.73e-27 | 47 | 381 | 38 | 391 | Chondroitinase-B OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=cslB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
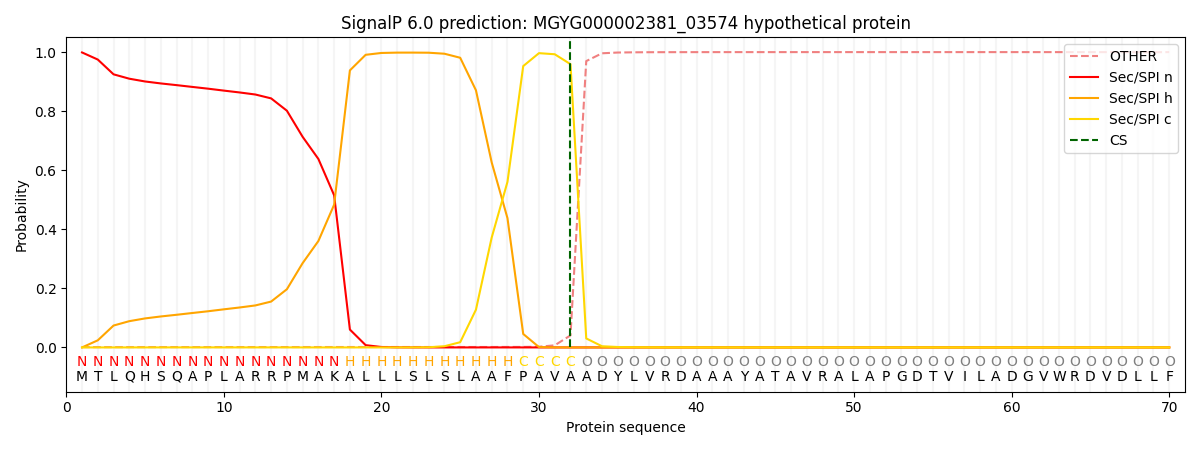
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002562 | 0.996309 | 0.000318 | 0.000326 | 0.000237 | 0.000213 |
