You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002382_00351
You are here: Home > Sequence: MGYG000002382_00351
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stenotrophomonas maltophilia_AK | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Stenotrophomonas; Stenotrophomonas maltophilia_AK | |||||||||||
| CAZyme ID | MGYG000002382_00351 | |||||||||||
| CAZy Family | PL5 | |||||||||||
| CAZyme Description | Polysaccharide lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5765; End: 6766 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL5 | 22 | 299 | 2.4e-99 | 0.9053627760252366 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK00325 | algL | 5.82e-127 | 23 | 326 | 49 | 359 | polysaccharide lyase. |
| cd00244 | AlgLyase | 5.89e-60 | 23 | 322 | 30 | 339 | Alginate Lyase A1-III; enzymatically depolymerizes alginate, a complex copolymer of beta-D-mannuronate and alpha-L-guluronate, by cleaving the beta-(1,4) glycosidic bond. |
| pfam05426 | Alginate_lyase | 5.17e-50 | 33 | 270 | 1 | 274 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYA90081.1 | 4.77e-247 | 1 | 332 | 1 | 332 |
| QNG81436.1 | 5.68e-232 | 1 | 332 | 1 | 331 |
| QJC75652.1 | 1.15e-231 | 1 | 332 | 1 | 331 |
| CAQ45011.1 | 2.31e-231 | 1 | 332 | 1 | 331 |
| ASE52599.1 | 2.31e-231 | 1 | 332 | 1 | 331 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7FHU_A | 8.21e-225 | 23 | 332 | 1 | 310 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHU_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHV_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHV_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHW_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHW_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHX_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHX_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHY_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHY_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FI0_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FI1_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 7FHZ_A | 3.66e-224 | 23 | 331 | 1 | 309 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 7FI2_A | 2.74e-223 | 23 | 332 | 1 | 310 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 4OZV_A | 3.43e-38 | 77 | 319 | 79 | 331 | CrystalStructure of the periplasmic alginate lyase AlgL [Pseudomonas aeruginosa PAO1] |
| 4OZW_A | 9.72e-37 | 77 | 319 | 79 | 331 | CrystalStructure of the periplasmic alginate lyase AlgL H202A mutant [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2FHL8 | 4.63e-232 | 1 | 332 | 1 | 331 | Polysaccharide lyase OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=Smlt1473 PE=1 SV=1 |
| C3KDZ3 | 6.62e-48 | 23 | 319 | 53 | 359 | Alginate lyase OS=Pseudomonas fluorescens (strain SBW25) OX=216595 GN=algL PE=3 SV=1 |
| P59786 | 6.62e-48 | 23 | 319 | 53 | 359 | Alginate lyase OS=Pseudomonas fluorescens OX=294 GN=algL PE=3 SV=1 |
| Q4KHY5 | 9.47e-47 | 23 | 319 | 54 | 360 | Alginate lyase OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=algL PE=3 SV=1 |
| Q3KHR0 | 1.99e-46 | 23 | 319 | 54 | 360 | Alginate lyase OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=algL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
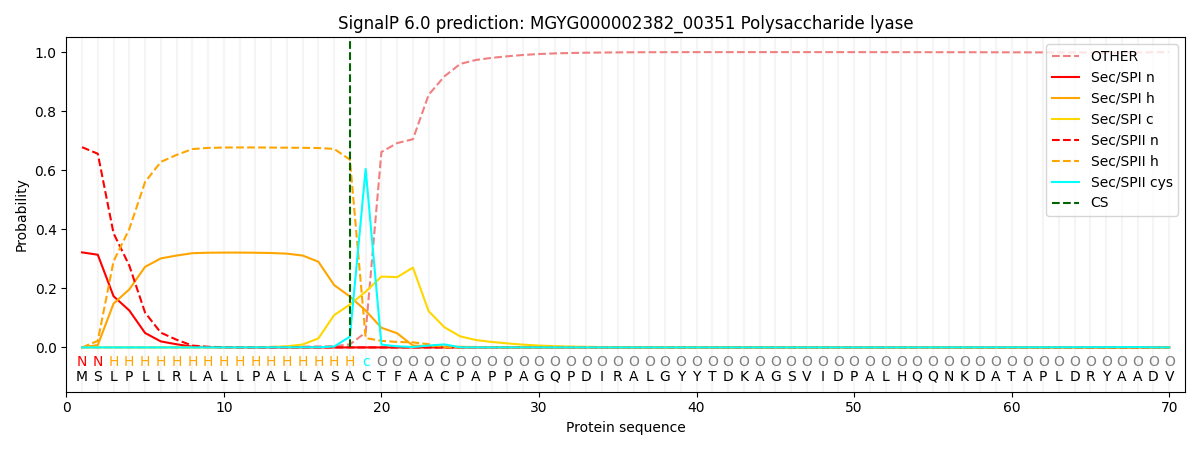
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000481 | 0.315660 | 0.683406 | 0.000161 | 0.000146 | 0.000132 |
