You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002383_00619
You are here: Home > Sequence: MGYG000002383_00619
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
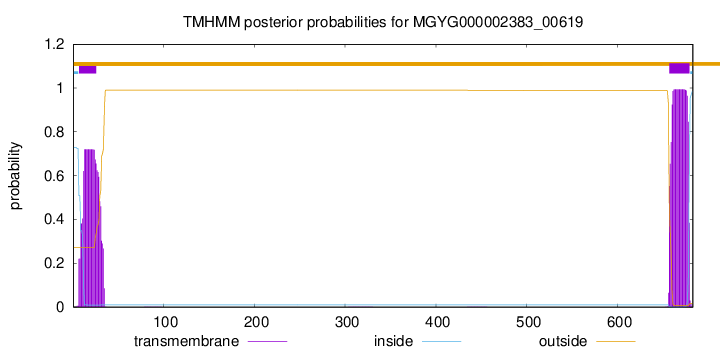
TMHMM annotations
Basic Information help
| Species | Lacticaseibacillus zeae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lacticaseibacillus; Lacticaseibacillus zeae | |||||||||||
| CAZyme ID | MGYG000002383_00619 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 134690; End: 136741 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 115 | 345 | 6.6e-55 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 2.36e-67 | 52 | 380 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 1.15e-58 | 51 | 387 | 1 | 318 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 4.58e-32 | 120 | 319 | 56 | 256 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 8.09e-22 | 41 | 386 | 35 | 358 | beta-glucosidase BglX. |
| pfam07554 | FIVAR | 8.17e-06 | 489 | 550 | 1 | 69 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVI32968.1 | 0.0 | 1 | 683 | 1 | 683 |
| ARY91138.1 | 0.0 | 1 | 683 | 1 | 683 |
| QPC16370.1 | 0.0 | 1 | 683 | 1 | 683 |
| BAN74096.1 | 0.0 | 1 | 683 | 1 | 683 |
| QXG59952.1 | 0.0 | 1 | 683 | 1 | 683 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BMX_A | 5.08e-46 | 43 | 399 | 34 | 411 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 1.72e-45 | 43 | 399 | 8 | 385 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 2.54e-45 | 43 | 399 | 38 | 415 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 6K5J_A | 4.69e-45 | 51 | 383 | 11 | 339 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3SQL_A | 4.88e-37 | 38 | 384 | 11 | 349 | CrystalStructure of Glycoside Hydrolase from Synechococcus [Synechococcus sp. PCC 7002],3SQL_B Crystal Structure of Glycoside Hydrolase from Synechococcus [Synechococcus sp. PCC 7002],3SQM_A Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_B Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_C Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002],3SQM_D Crystal Structure of Glycoside Hydrolase from Synechococcus Complexed with N-acetyl-D-glucosamine [Synechococcus sp. PCC 7002] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 2.78e-45 | 43 | 399 | 34 | 411 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| Q5H1Q0 | 2.65e-32 | 69 | 337 | 12 | 277 | Beta-hexosaminidase OS=Xanthomonas oryzae pv. oryzae (strain KACC10331 / KXO85) OX=291331 GN=nagZ PE=3 SV=2 |
| Q2P4L0 | 9.08e-32 | 69 | 337 | 12 | 277 | Beta-hexosaminidase OS=Xanthomonas oryzae pv. oryzae (strain MAFF 311018) OX=342109 GN=nagZ PE=3 SV=1 |
| Q4USG7 | 7.34e-31 | 69 | 337 | 12 | 277 | Beta-hexosaminidase OS=Xanthomonas campestris pv. campestris (strain 8004) OX=314565 GN=nagZ PE=3 SV=1 |
| B0RX17 | 7.34e-31 | 69 | 337 | 12 | 277 | Beta-hexosaminidase OS=Xanthomonas campestris pv. campestris (strain B100) OX=509169 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
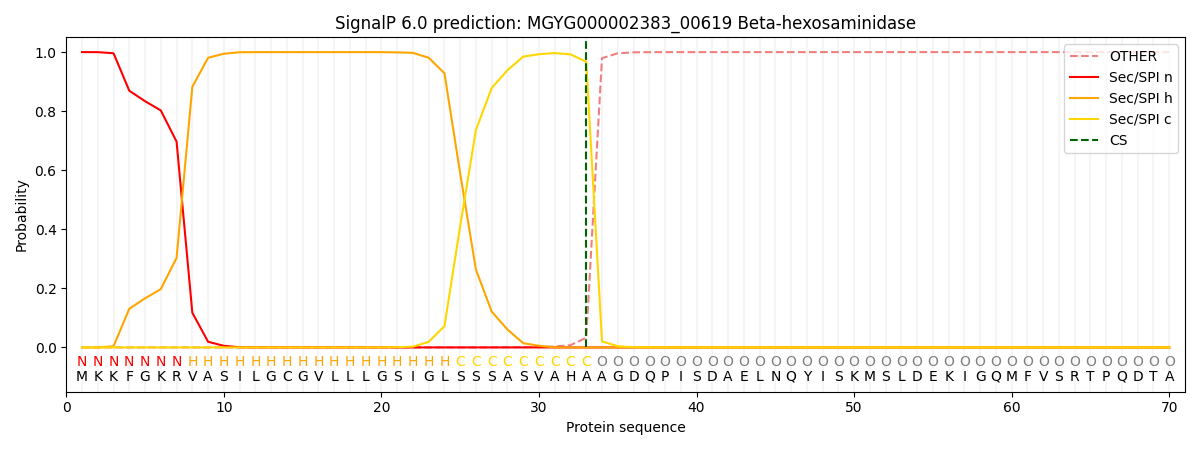
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999013 | 0.000205 | 0.000181 | 0.000162 | 0.000142 |