You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002386_02906
You are here: Home > Sequence: MGYG000002386_02906
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lactiplantibacillus plantarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lactiplantibacillus; Lactiplantibacillus plantarum | |||||||||||
| CAZyme ID | MGYG000002386_02906 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3005633; End: 3006700 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 1.27e-26 | 261 | 349 | 1 | 98 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 8.62e-19 | 252 | 351 | 78 | 193 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| NF033742 | NlpC_p60_RipB | 3.08e-15 | 252 | 349 | 80 | 198 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
| PRK13914 | PRK13914 | 5.73e-13 | 252 | 330 | 369 | 454 | invasion associated endopeptidase. |
| PRK10838 | spr | 3.77e-12 | 261 | 351 | 86 | 178 | bifunctional murein DD-endopeptidase/murein LD-carboxypeptidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMX11635.1 | 6.64e-110 | 1 | 355 | 1 | 355 |
| QQM60794.1 | 6.64e-110 | 1 | 355 | 1 | 355 |
| QSW66663.1 | 6.64e-110 | 1 | 355 | 1 | 355 |
| QHM46220.1 | 6.64e-110 | 1 | 355 | 1 | 355 |
| AHN70445.1 | 6.64e-110 | 1 | 355 | 1 | 355 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B8C_A | 5.02e-25 | 252 | 355 | 31 | 142 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 2EVR_A | 2.51e-08 | 261 | 332 | 130 | 201 | ChainA, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102],2FG0_A Chain A, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102],2FG0_B Chain B, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102] |
| 2HBW_A | 1.35e-07 | 261 | 332 | 119 | 190 | ChainA, NLP/P60 protein [Trichormus variabilis ATCC 29413] |
| 4XCM_A | 2.09e-06 | 252 | 343 | 117 | 214 | Crystalstructure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8],4XCM_B Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus [Thermus thermophilus HB8] |
| 4Q4T_A | 4.25e-06 | 268 | 349 | 382 | 464 | Structureof the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13692 | 7.48e-23 | 252 | 355 | 405 | 516 | Protein P54 OS=Enterococcus faecium OX=1352 PE=3 SV=2 |
| Q8NNK6 | 8.44e-11 | 252 | 346 | 101 | 200 | Probable endopeptidase Cgl2188 OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=Cgl2188 PE=3 SV=1 |
| A4QFQ3 | 8.70e-11 | 252 | 346 | 103 | 202 | Probable endopeptidase cgR_2070 OS=Corynebacterium glutamicum (strain R) OX=340322 GN=cgR_2070 PE=1 SV=1 |
| Q01835 | 4.79e-10 | 254 | 339 | 401 | 493 | Probable endopeptidase p60 OS=Listeria grayi OX=1641 GN=iap PE=3 SV=1 |
| P21171 | 1.45e-09 | 252 | 330 | 372 | 457 | Probable endopeptidase p60 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=iap PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
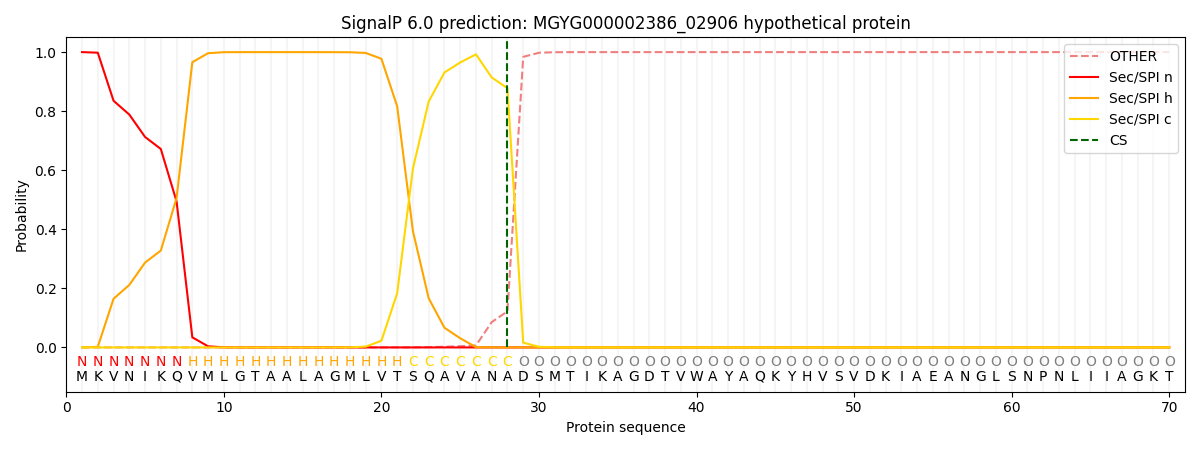
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000225 | 0.999102 | 0.000166 | 0.000185 | 0.000161 | 0.000141 |
