You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002389_01991
You are here: Home > Sequence: MGYG000002389_01991
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Latilactobacillus sakei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Latilactobacillus; Latilactobacillus sakei | |||||||||||
| CAZyme ID | MGYG000002389_01991 | |||||||||||
| CAZy Family | GH70 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2889; End: 7505 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH70 | 497 | 1293 | 0 | 0.9975093399750934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02324 | Glyco_hydro_70 | 0.0 | 497 | 1295 | 1 | 804 | Glycosyl hydrolase family 70. Members of this family belong to glycosyl hydrolase family 70 Glucosyltransferases or sucrose 6-glycosyl transferases (GTF-S) catalyze the transfer of D-glucopyramnosyl units from sucrose onto acceptor molecules, EC:2.4.1.5. This family roughly corresponds to the N-terminal catalytic domain of the enzyme. Members of this family also contain the Putative cell wall binding domain pfam01473, which corresponds with the C-terminal glucan-binding domain. |
| TIGR04035 | glucan_65_rpt | 9.11e-18 | 1320 | 1381 | 1 | 62 | glucan-binding repeat. This model describes a region of about 63 amino acids that is composed of three repeats of a more broadly distributed family of shorter repeats modeled by pfam01473. While the shorter repeats are often associated with choline binding (and therefore with cell wall binding), the longer repeat described here represents a subgroup of repeat sequences associated with glucan binding, as found in a number glycosylhydrolases. Shah, et al. describe a repeat consensus, WYYFDANGKAVTGAQTINGQTLYFDQDGKQVKG, that corresponds to half of the repeat as modeled here and one and a half copies of the repeat as modeled by pfam01473. |
| COG5263 | COG5263 | 6.44e-17 | 1299 | 1473 | 142 | 309 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| COG5263 | COG5263 | 7.24e-15 | 285 | 426 | 132 | 272 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| COG5263 | COG5263 | 1.77e-13 | 1227 | 1476 | 52 | 292 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SON74263.1 | 0.0 | 1 | 1497 | 1 | 1497 |
| ATN28243.1 | 0.0 | 1 | 1497 | 1 | 1497 |
| AXN36915.1 | 0.0 | 1 | 1485 | 1 | 1485 |
| CCK33643.1 | 0.0 | 1 | 1485 | 1 | 1485 |
| AAU08011.1 | 0.0 | 1 | 1485 | 1 | 1458 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5NGY_A | 0.0 | 277 | 1392 | 8 | 1268 | Crystalstructure of Leuconostoc citreum NRRL B-1299 dextransucrase DSR-M [Leuconostoc citreum],5NGY_B Crystal structure of Leuconostoc citreum NRRL B-1299 dextransucrase DSR-M [Leuconostoc citreum],5O8L_A Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with sucrose [Leuconostoc citreum],6HTV_A Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M in complex with isomaltotetraose [Leuconostoc citreum] |
| 5LFC_A | 0.0 | 277 | 1392 | 11 | 1271 | Crystalstructure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M [Leuconostoc citreum],5LFC_B Crystal structure of Leuconostoc citreum NRRL B-1299 N-terminally truncated dextransucrase DSR-M [Leuconostoc citreum] |
| 6SYQ_B | 1.22e-316 | 284 | 1353 | 3 | 1145 | ChainB, Alternansucrase [Leuconostoc mesenteroides] |
| 6SYQ_A | 7.06e-307 | 415 | 1353 | 222 | 1234 | ChainA, Alternansucrase [Leuconostoc mesenteroides] |
| 6SZI_A | 7.76e-307 | 415 | 1353 | 225 | 1237 | ChainA, Alternansucrase [Leuconostoc mesenteroides],6SZI_B Chain B, Alternansucrase [Leuconostoc mesenteroides],6T16_A Chain A, Alternansucrase [Leuconostoc mesenteroides],6T16_B Chain B, Alternansucrase [Leuconostoc mesenteroides],6T18_A ASR Alternansucrase in complex with oligoalternan [Leuconostoc mesenteroides],6T18_B ASR Alternansucrase in complex with oligoalternan [Leuconostoc mesenteroides],6T1P_A Chain A, Alternansucrase [Leuconostoc mesenteroides],6T1P_B Chain B, Alternansucrase [Leuconostoc mesenteroides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P08987 | 0.0 | 375 | 1480 | 167 | 1256 | Glucosyltransferase-I OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=gtfB PE=3 SV=3 |
| P13470 | 0.0 | 450 | 1480 | 248 | 1284 | Glucosyltransferase-SI OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=gtfC PE=1 SV=2 |
| P49331 | 0.0 | 414 | 1480 | 198 | 1288 | Glucosyltransferase-S OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=gtfD PE=3 SV=3 |
| P11001 | 1.75e-315 | 437 | 1480 | 207 | 1258 | Glucosyltransferase-I OS=Streptococcus downei OX=1317 GN=gtfI PE=3 SV=1 |
| P27470 | 5.95e-315 | 450 | 1480 | 216 | 1253 | Glucosyltransferase-I OS=Streptococcus downei OX=1317 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
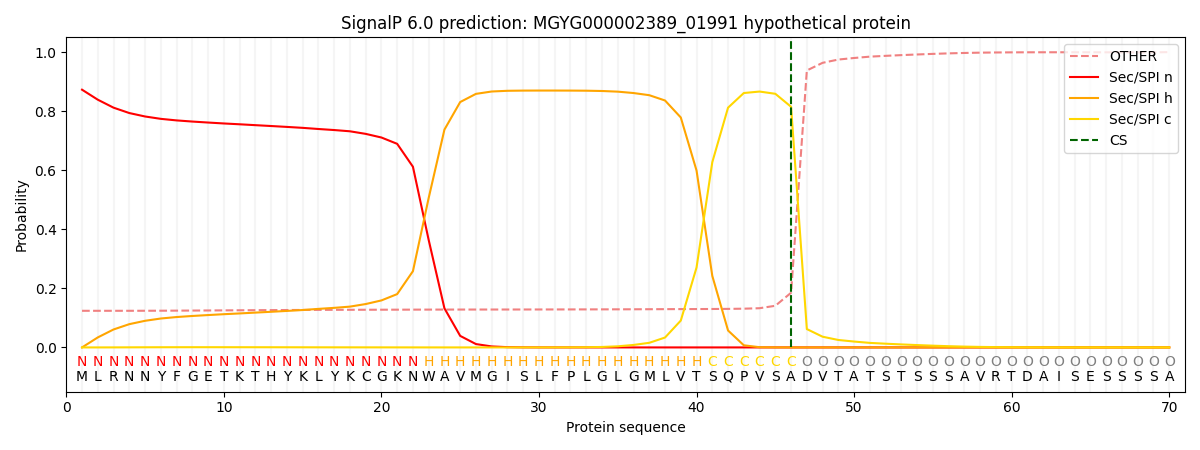
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.132232 | 0.862208 | 0.004548 | 0.000362 | 0.000279 | 0.000361 |
