You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002391_02370
You are here: Home > Sequence: MGYG000002391_02370
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
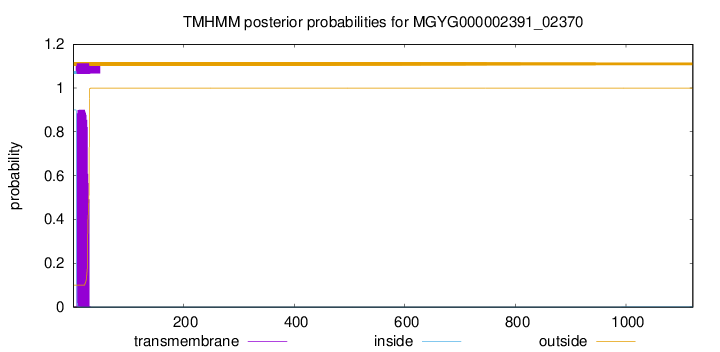
TMHMM annotations
Basic Information help
| Species | Oceanobacillus caeni | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Oceanobacillus; Oceanobacillus caeni | |||||||||||
| CAZyme ID | MGYG000002391_02370 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10625; End: 13990 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 885 | 1029 | 1.8e-18 | 0.953125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.80e-59 | 802 | 1047 | 24 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 4.15e-26 | 873 | 1038 | 1 | 146 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam08239 | SH3_3 | 1.32e-10 | 1064 | 1117 | 1 | 53 | Bacterial SH3 domain. |
| COG4991 | YraI | 7.43e-10 | 1057 | 1117 | 30 | 92 | Uncharacterized conserved protein YraI [Function unknown]. |
| smart00287 | SH3b | 1.24e-07 | 1059 | 1115 | 4 | 59 | Bacterial SH3 domain homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SQI62956.1 | 2.25e-244 | 453 | 1117 | 40 | 722 |
| QHS23523.1 | 5.03e-241 | 447 | 1119 | 175 | 850 |
| QTN00811.1 | 6.91e-222 | 453 | 1117 | 248 | 912 |
| VEF49624.1 | 1.53e-209 | 452 | 1119 | 40 | 725 |
| QQZ10351.1 | 2.99e-182 | 453 | 1047 | 275 | 914 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 1.24e-43 | 832 | 1047 | 40 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 5.69e-32 | 809 | 1035 | 55 | 263 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 6FXP_A | 6.76e-32 | 809 | 1035 | 25 | 233 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 2.16e-31 | 855 | 1021 | 55 | 201 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 5WQW_A | 3.52e-31 | 781 | 1048 | 1 | 270 | X-raystructure of catalytic domain of autolysin from Clostridium perfringens [Clostridium perfringens str. 13] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 2.00e-50 | 804 | 1047 | 597 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| O33635 | 3.91e-41 | 832 | 1047 | 1131 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 3.91e-41 | 832 | 1047 | 1131 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q8CPQ1 | 6.78e-41 | 832 | 1045 | 1131 | 1332 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q2FZK7 | 5.12e-39 | 832 | 1047 | 1052 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000389 | 0.998926 | 0.000200 | 0.000165 | 0.000148 | 0.000147 |