You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002392_00052
You are here: Home > Sequence: MGYG000002392_00052
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
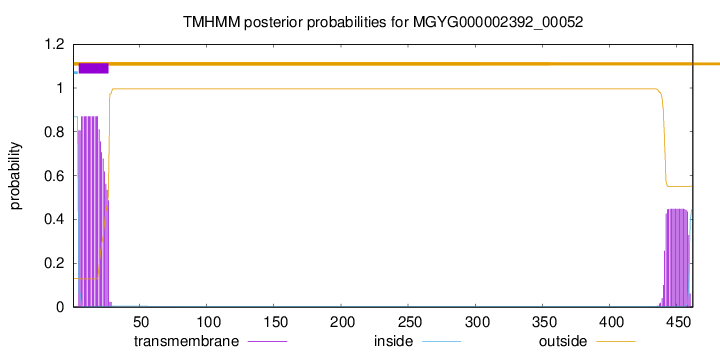
TMHMM annotations
Basic Information help
| Species | Listeria monocytogenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria monocytogenes | |||||||||||
| CAZyme ID | MGYG000002392_00052 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 63449; End: 64837 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4932 | COG4932 | 1.29e-79 | 1 | 311 | 635 | 965 | Uncharacterized surface anchored protein [Function unknown]. |
| pfam05737 | Collagen_bind | 8.60e-34 | 177 | 295 | 1 | 129 | Collagen binding domain. The domain fold is a jelly-roll, composed of two antiparallel beta-sheets and two short alpha-helices. A groove on beta-sheet I exhibited the best surface complementarity to the collagen. This site partially overlaps with the peptide sequence previously shown to be critical for collagen binding. Recombinant proteins containing single amino acid mutations designed to disrupt the surface of the putative binding site exhibited significantly lower affinities for collagen. |
| pfam17961 | Big_8 | 2.06e-19 | 49 | 144 | 1 | 100 | Bacterial Ig domain. This entry represents a bacterial Ig-fold domain that is found in a wide range of bacterial cell surface adherence proteins. |
| COG1388 | LysM | 3.69e-13 | 292 | 416 | 2 | 124 | LysM repeat [Cell wall/membrane/envelope biogenesis]. |
| pfam01476 | LysM | 3.97e-12 | 358 | 400 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBY54227.1 | 0.0 | 1 | 462 | 1 | 462 |
| CAC98958.1 | 0.0 | 1 | 462 | 1 | 462 |
| AEO25179.1 | 0.0 | 1 | 462 | 1 | 462 |
| CBY57161.1 | 0.0 | 1 | 462 | 1 | 462 |
| ANE38742.1 | 0.0 | 1 | 462 | 1 | 462 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2MKX_A | 3.60e-06 | 358 | 397 | 7 | 46 | Solutionstructure of LysM the peptidoglycan binding domain of autolysin AtlA from Enterococcus faecalis [Enterococcus faecalis V583] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8Y8L7 | 0.0 | 1 | 462 | 1 | 462 | Cell wall protein Lmo0880 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=lmo0880 PE=1 SV=1 |
| P11000 | 3.71e-17 | 1 | 291 | 3 | 316 | Wall-associated protein OS=Streptococcus mutans serotype c (strain ATCC 700610 / UA159) OX=210007 GN=wapA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
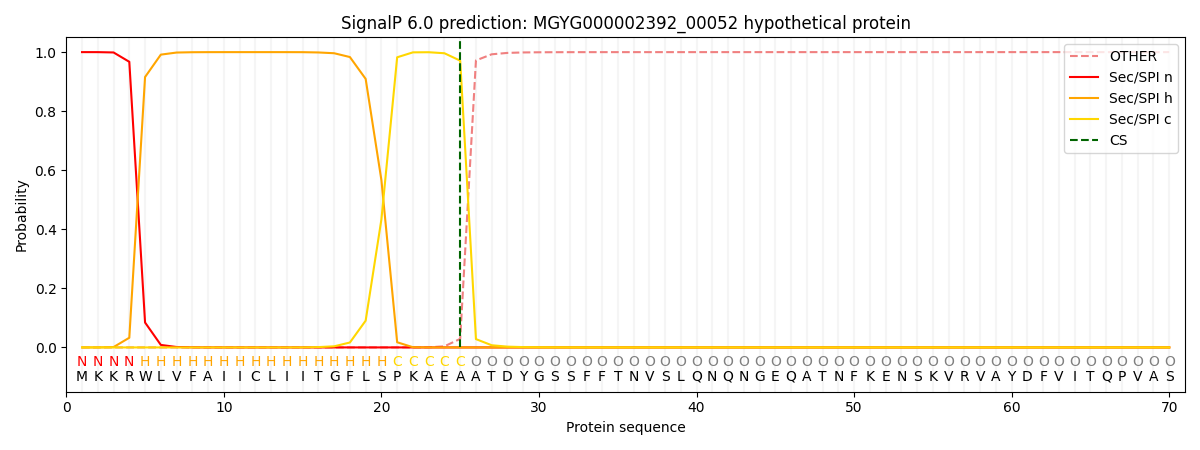
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000329 | 0.998547 | 0.000475 | 0.000205 | 0.000215 | 0.000184 |