You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002392_01375
You are here: Home > Sequence: MGYG000002392_01375
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Listeria monocytogenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria monocytogenes | |||||||||||
| CAZyme ID | MGYG000002392_01375 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 549742; End: 550770 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 23 | 328 | 7.8e-87 | 0.8715083798882681 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4833 | COG4833 | 0.0 | 1 | 341 | 1 | 353 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| pfam03663 | Glyco_hydro_76 | 1.06e-32 | 20 | 283 | 6 | 299 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG1331 | YyaL | 0.001 | 49 | 254 | 212 | 452 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| pfam00759 | Glyco_hydro_9 | 0.006 | 88 | 135 | 209 | 257 | Glycosyl hydrolase family 9. |
| COG1331 | YyaL | 0.009 | 123 | 277 | 406 | 517 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBY51238.1 | 3.29e-261 | 1 | 342 | 1 | 342 |
| CDG44635.1 | 3.29e-261 | 1 | 342 | 1 | 342 |
| AMD51478.1 | 3.29e-261 | 1 | 342 | 1 | 342 |
| QOF60393.1 | 3.29e-261 | 1 | 342 | 1 | 342 |
| AKI54806.1 | 3.29e-261 | 1 | 342 | 1 | 342 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3K7X_A | 3.21e-246 | 2 | 341 | 2 | 341 | ChainA, Lin0763 protein [Listeria innocua] |
| 6SHD_A | 2.76e-63 | 3 | 341 | 54 | 390 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 8.68e-62 | 3 | 341 | 55 | 391 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 6.75e-61 | 3 | 341 | 55 | 391 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 4BOK_A | 1.27e-25 | 44 | 286 | 37 | 282 | ChainA, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05031 | 1.66e-07 | 109 | 317 | 145 | 370 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
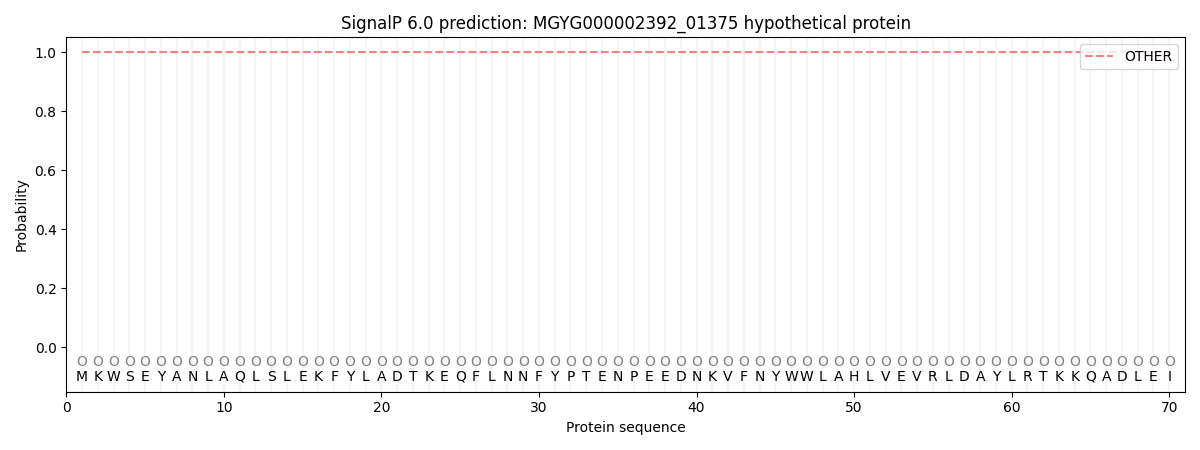
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000046 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
