You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002395_00139
You are here: Home > Sequence: MGYG000002395_00139
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
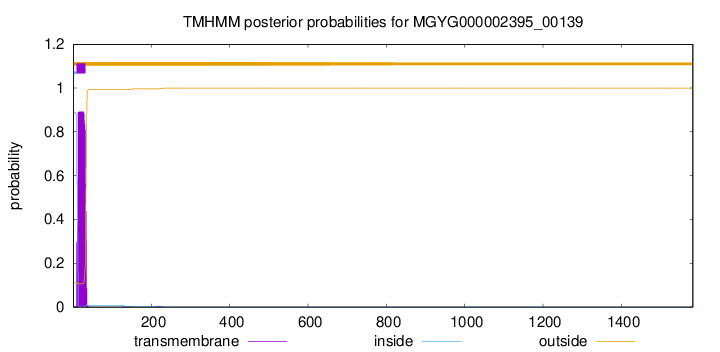
TMHMM annotations
Basic Information help
| Species | Bifidobacterium adolescentis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium adolescentis | |||||||||||
| CAZyme ID | MGYG000002395_00139 | |||||||||||
| CAZy Family | GH121 | |||||||||||
| CAZyme Description | Beta-L-arabinobiosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 184882; End: 189633 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH121 | 13 | 1444 | 0 | 0.992816091954023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07532 | Big_4 | 7.67e-07 | 1287 | 1347 | 1 | 58 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07532 | Big_4 | 8.13e-07 | 1371 | 1431 | 2 | 59 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJE05111.1 | 0.0 | 1 | 1583 | 1 | 1583 |
| QHB61921.1 | 0.0 | 1 | 1579 | 1 | 1579 |
| AZN74101.1 | 0.0 | 1 | 1579 | 1 | 1579 |
| QTL81705.1 | 0.0 | 1 | 1579 | 1 | 1579 |
| BAR02807.1 | 0.0 | 1 | 1579 | 1 | 1579 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M5A_A | 0.0 | 35 | 880 | 3 | 865 | Crystalstructure of GH121 beta-L-arabinobiosidase HypBA2 from Bifidobacterium longum [Bifidobacterium longum] |
| 5MQS_A | 8.33e-11 | 328 | 831 | 651 | 1106 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 5MQR_A | 1.87e-08 | 328 | 831 | 651 | 1106 | SialidaseBT_1020 [Bacteroides thetaiotaomicron] |
| 7NIT_A | 9.86e-08 | 1291 | 1441 | 896 | 1035 | ChainA, Beta-galactosidase [Bifidobacterium bifidum],7NIT_B Chain B, Beta-galactosidase [Bifidobacterium bifidum],7NIT_C Chain C, Beta-galactosidase [Bifidobacterium bifidum],7NIT_D Chain D, Beta-galactosidase [Bifidobacterium bifidum],7NIT_E Chain E, Beta-galactosidase [Bifidobacterium bifidum],7NIT_F Chain F, Beta-galactosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 0.0 | 4 | 1583 | 4 | 1598 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
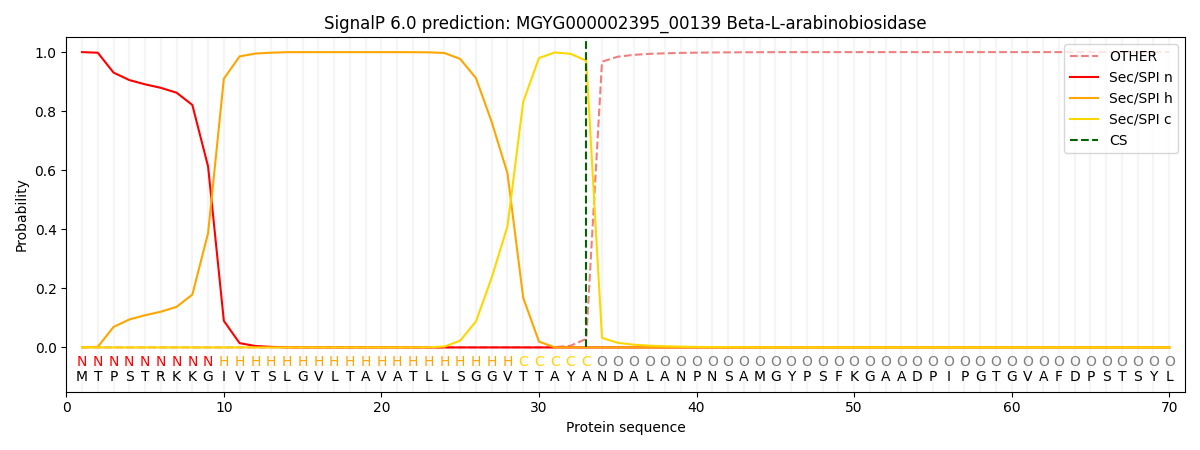
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000493 | 0.998546 | 0.000238 | 0.000278 | 0.000204 | 0.000175 |