You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002396_00526
You are here: Home > Sequence: MGYG000002396_00526
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
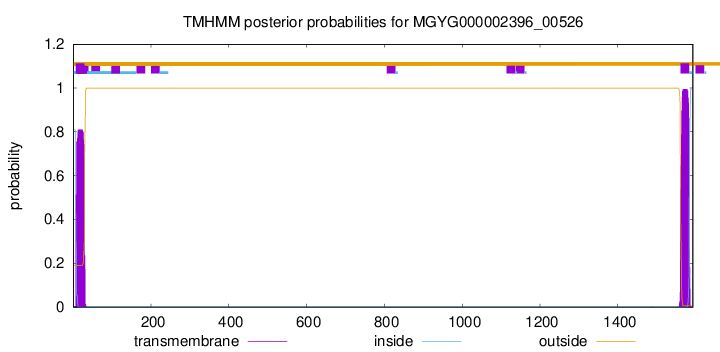
TMHMM annotations
Basic Information help
| Species | Bifidobacterium bifidum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium bifidum | |||||||||||
| CAZyme ID | MGYG000002396_00526 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 645869; End: 650650 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033681 | ExeM_NucH_DNase | 0.0 | 269 | 875 | 7 | 544 | ExeM/NucH family extracellular endonuclease. |
| COG2374 | COG2374 | 1.77e-116 | 9 | 882 | 4 | 794 | Predicted extracellular nuclease [General function prediction only]. |
| PRK09419 | PRK09419 | 6.06e-65 | 881 | 1419 | 660 | 1146 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| COG0737 | UshA | 4.61e-64 | 879 | 1408 | 24 | 501 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| NF033680 | exonuc_ExeM-GG | 1.37e-61 | 44 | 875 | 26 | 844 | extracellular exonuclease ExeM. ExeM, as described in Shewanella oneidensis, is a biofilm formation-associated exonuclease that cleaves extracellular DNA (eDNA), a biofilm component. Members of the ExeM family contain two or three pairs of Cys residues, presumed to form disulfide bonds, and a C-terminal GlyGly-CTERM membrane-anchoring segment. Strangely, engineered removal of the GlyGly-CTERM region did not result in net export from the cell and appearance of the enzyme in culture supernatants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDS92078.1 | 0.0 | 44 | 1510 | 44 | 1466 |
| QAY73698.1 | 0.0 | 42 | 1516 | 83 | 1529 |
| ACZ32093.1 | 0.0 | 13 | 1510 | 12 | 1469 |
| QAY64865.1 | 0.0 | 14 | 1510 | 14 | 1477 |
| QAY71724.1 | 0.0 | 44 | 1466 | 40 | 1430 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3IVD_A | 4.04e-27 | 933 | 1391 | 69 | 469 | Putative5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine [Escherichia coli O6],3IVD_B Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine [Escherichia coli O6],3IVE_A Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine [Escherichia coli O6] |
| 1OI8_A | 2.11e-24 | 879 | 1408 | 6 | 511 | 5'-Nucleotidase(E. coli) with an Engineered Disulfide Bridge (P90C, L424C) [Escherichia coli],1OI8_B 5'-Nucleotidase (E. coli) with an Engineered Disulfide Bridge (P90C, L424C) [Escherichia coli] |
| 1HP1_A | 3.27e-24 | 879 | 1408 | 6 | 502 | 5'-Nucleotidase(Open Form) Complex With Atp [Escherichia coli] |
| 1HO5_A | 3.53e-24 | 879 | 1408 | 6 | 511 | 5'-Nucleotidase(E. Coli) In Complex With Adenosine And Phosphate [Escherichia coli],1HO5_B 5'-Nucleotidase (E. Coli) In Complex With Adenosine And Phosphate [Escherichia coli],1HPU_A 5'-Nucleotidase (Closed Form), Complex With Ampcp [Escherichia coli],1HPU_B 5'-Nucleotidase (Closed Form), Complex With Ampcp [Escherichia coli],1HPU_C 5'-Nucleotidase (Closed Form), Complex With Ampcp [Escherichia coli],1HPU_D 5'-Nucleotidase (Closed Form), Complex With Ampcp [Escherichia coli] |
| 1USH_A | 4.27e-24 | 879 | 1408 | 31 | 536 | 5'-NucleotidaseFrom E. Coli [Escherichia coli K-12],2USH_A 5'-Nucleotidase From E. Coli [Escherichia coli K-12],2USH_B 5'-Nucleotidase From E. Coli [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54602 | 6.35e-37 | 882 | 1402 | 590 | 1068 | Endonuclease YhcR OS=Bacillus subtilis (strain 168) OX=224308 GN=yhcR PE=1 SV=1 |
| Q8YAJ5 | 2.04e-33 | 875 | 1402 | 34 | 562 | Cell wall protein Lmo0130 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=lmo0130 PE=1 SV=1 |
| A9BJC1 | 1.07e-29 | 877 | 1403 | 19 | 482 | Mannosylglucosyl-3-phosphoglycerate phosphatase OS=Petrotoga mobilis (strain DSM 10674 / SJ95) OX=403833 GN=mggB PE=1 SV=1 |
| Q8DFG4 | 4.52e-28 | 875 | 1408 | 31 | 539 | 5'-nucleotidase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=nutA PE=3 SV=2 |
| Q9KQ30 | 6.02e-27 | 876 | 1408 | 32 | 539 | 5'-nucleotidase OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=nutA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
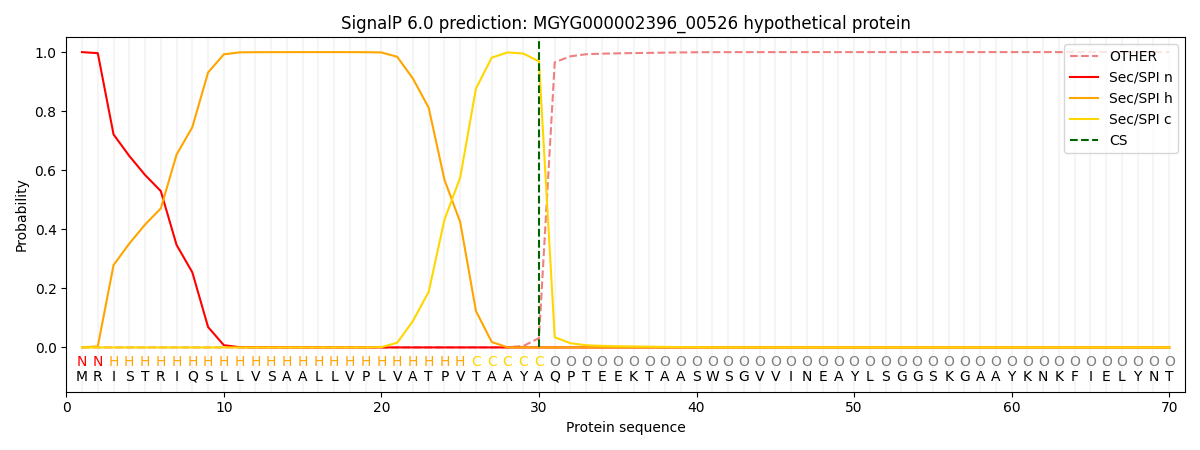
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000447 | 0.998658 | 0.000183 | 0.000287 | 0.000216 | 0.000182 |