You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002401_00306
You are here: Home > Sequence: MGYG000002401_00306
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
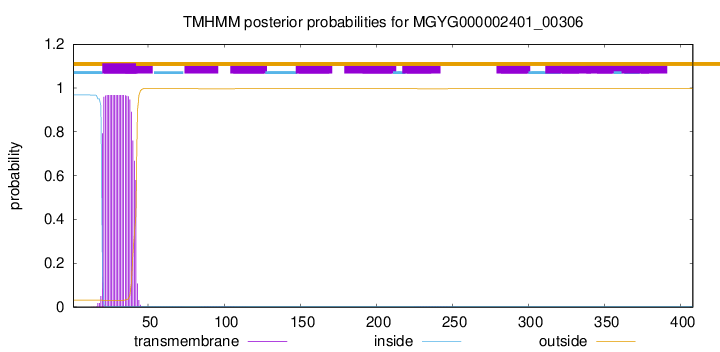
TMHMM annotations
Basic Information help
| Species | Propionibacterium freudenreichii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Propionibacterium; Propionibacterium freudenreichii | |||||||||||
| CAZyme ID | MGYG000002401_00306 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 360135; End: 361361 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 139 | 359 | 1e-33 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 2.55e-39 | 105 | 403 | 25 | 317 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 9.05e-29 | 105 | 397 | 27 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 1.73e-24 | 110 | 359 | 26 | 278 | beta-hexosaminidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL57497.1 | 6.15e-255 | 1 | 408 | 6 | 413 |
| AJQ91615.1 | 3.06e-254 | 1 | 408 | 2 | 409 |
| CEP26529.1 | 1.45e-253 | 1 | 408 | 6 | 413 |
| CUW07176.1 | 2.06e-252 | 1 | 408 | 2 | 409 |
| CUW21858.1 | 2.06e-252 | 1 | 408 | 2 | 409 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BU9_A | 5.21e-54 | 86 | 395 | 13 | 334 | Crystalstructure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333],5BU9_B Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 [Beutenbergia cavernae DSM 12333] |
| 3TEV_A | 1.15e-22 | 102 | 403 | 36 | 347 | Thecrystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1],3TEV_B The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 [Deinococcus radiodurans R1] |
| 6K5J_A | 9.16e-21 | 139 | 359 | 78 | 297 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 5G1M_A | 4.11e-20 | 127 | 359 | 65 | 293 | Crystalstructure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G1M_B Crystal structure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G2M_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G2M_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G3R_A Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G3R_B Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G5K_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5G5K_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
| 5G5U_A | 5.58e-20 | 127 | 359 | 65 | 293 | Crystalstructure of NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G5U_B Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G6T_A Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G6T_B Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5LY7_A Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5LY7_B Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5H1Q0 | 1.59e-23 | 111 | 363 | 26 | 280 | Beta-hexosaminidase OS=Xanthomonas oryzae pv. oryzae (strain KACC10331 / KXO85) OX=291331 GN=nagZ PE=3 SV=2 |
| B4SRK3 | 5.67e-23 | 111 | 363 | 26 | 280 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain R551-3) OX=391008 GN=nagZ PE=3 SV=1 |
| B2FPW9 | 7.76e-23 | 111 | 363 | 26 | 280 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=nagZ PE=3 SV=1 |
| Q2P4L0 | 1.43e-22 | 111 | 363 | 26 | 280 | Beta-hexosaminidase OS=Xanthomonas oryzae pv. oryzae (strain MAFF 311018) OX=342109 GN=nagZ PE=3 SV=1 |
| B0RX17 | 6.55e-22 | 111 | 363 | 26 | 280 | Beta-hexosaminidase OS=Xanthomonas campestris pv. campestris (strain B100) OX=509169 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000376 | 0.013086 | 0.986525 | 0.000040 | 0.000010 | 0.000004 |