You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002402_02269
You are here: Home > Sequence: MGYG000002402_02269
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
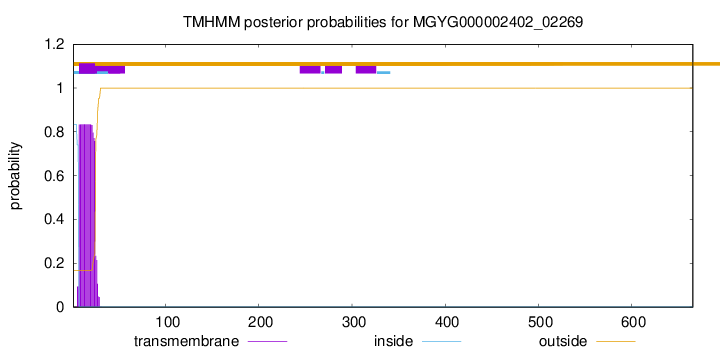
TMHMM annotations
Basic Information help
| Species | Clostridium_H massiliodielmoense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_H; Clostridium_H massiliodielmoense | |||||||||||
| CAZyme ID | MGYG000002402_02269 | |||||||||||
| CAZy Family | GH156 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1187355; End: 1189355 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH156 | 370 | 662 | 1.3e-22 | 0.5380434782608695 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02638 | GHL10 | 6.59e-04 | 348 | 542 | 31 | 260 | Glycosyl hydrolase-like 10. This is family of bacterial glycosyl-hydrolase-like proteins falling into the family GHL10 as described above,. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABK62041.1 | 0.0 | 9 | 666 | 1 | 656 |
| QPW57243.1 | 0.0 | 1 | 663 | 1 | 661 |
| AEB75368.1 | 0.0 | 25 | 663 | 15 | 651 |
| QPW60018.1 | 0.0 | 1 | 663 | 1 | 661 |
| AYF53429.1 | 0.0 | 1 | 663 | 1 | 661 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OQ2_A | 6.29e-07 | 445 | 636 | 158 | 381 | ChainA, Cwp19 [Clostridioides difficile],5OQ2_B Chain B, Cwp19 [Clostridioides difficile] |
| 5OQ3_A | 6.29e-07 | 445 | 636 | 158 | 381 | ChainA, Cwp19 [Clostridioides difficile] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
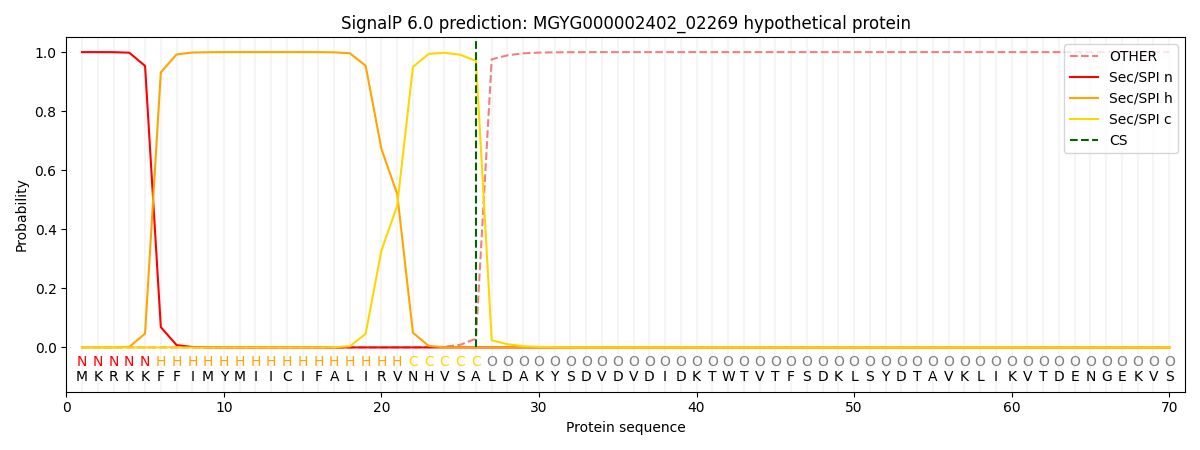
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000304 | 0.998974 | 0.000244 | 0.000151 | 0.000154 | 0.000146 |