You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002403_03524
You are here: Home > Sequence: MGYG000002403_03524
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
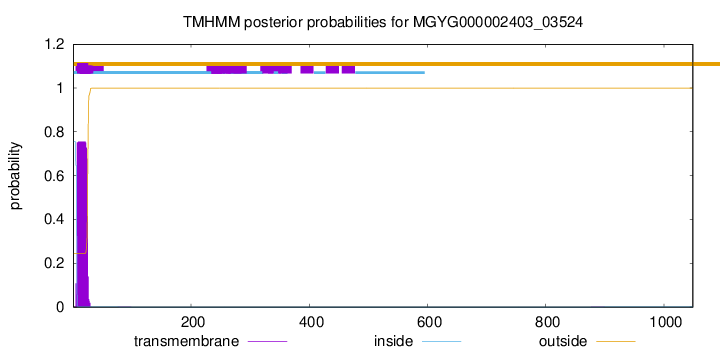
TMHMM annotations
Basic Information help
| Species | Robinsoniella peoriensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella peoriensis | |||||||||||
| CAZyme ID | MGYG000002403_03524 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20563; End: 23712 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 205 | 323 | 2.5e-20 | 0.9032258064516129 |
| CBM32 | 497 | 598 | 1.1e-16 | 0.8145161290322581 |
| CBM32 | 339 | 462 | 7.6e-16 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.30e-20 | 204 | 322 | 5 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02585 | PIG-L | 1.56e-14 | 624 | 696 | 1 | 79 | GlcNAc-PI de-N-acetylase. Members of this family are related to PIG-L an N-acetylglucosaminylphosphatidylinositol de-N-acetylase (EC:3.5.1.89) that catalyzes the second step in GPI biosynthesis. |
| pfam00754 | F5_F8_type_C | 5.43e-13 | 340 | 468 | 6 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 9.20e-12 | 498 | 596 | 6 | 113 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG2120 | LmbE | 6.19e-10 | 622 | 696 | 13 | 89 | N-acetylglucosaminyl deacetylase, LmbE family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO38183.1 | 0.0 | 61 | 1048 | 36 | 1021 |
| QBF74995.1 | 0.0 | 61 | 1048 | 36 | 1021 |
| QMW76217.1 | 5.49e-51 | 64 | 574 | 561 | 988 |
| QIB55915.1 | 5.49e-51 | 64 | 574 | 561 | 988 |
| QRO38181.1 | 1.66e-50 | 64 | 596 | 567 | 1243 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
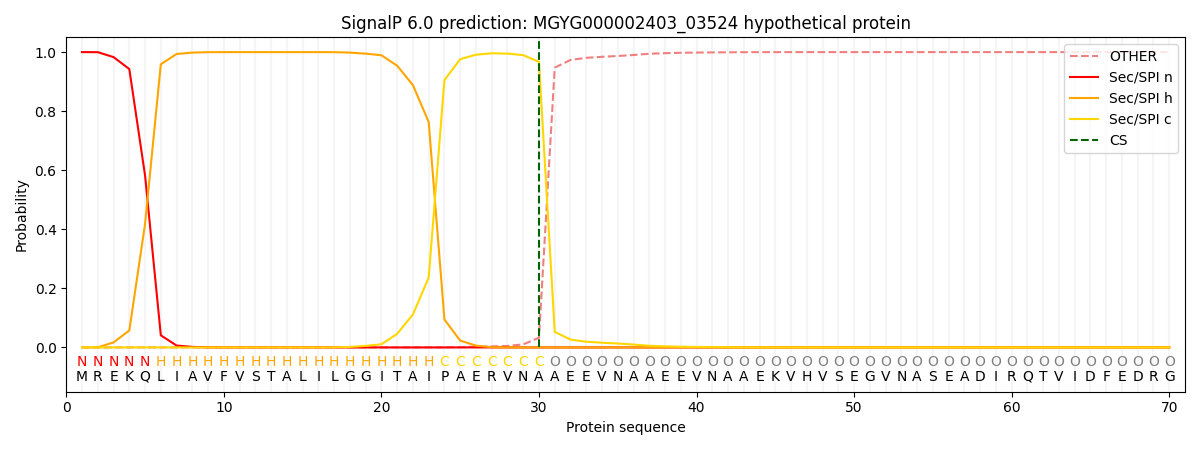
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000243 | 0.999065 | 0.000186 | 0.000201 | 0.000161 | 0.000140 |