You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002404_00142
You are here: Home > Sequence: MGYG000002404_00142
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
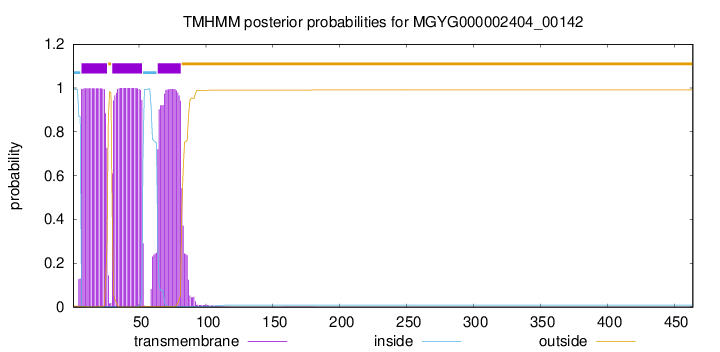
TMHMM annotations
Basic Information help
| Species | Bacillus_BE massilionigeriensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Bacillus_BE; Bacillus_BE massilionigeriensis | |||||||||||
| CAZyme ID | MGYG000002404_00142 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 142640; End: 144034 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05105 | Phage_holin_4_1 | 8.64e-46 | 29 | 132 | 6 | 109 | Bacteriophage holin family. Phage holins and lytic enzymes are both necessary for bacterial lysis and virus dissemination. This family also includes TcdE/UtxA involved in toxin secretion in Clostridium difficile. The 1.E.10 family is represented by Bacillus subtilis phi29 holin; 1.E.16 represents the Cph1 holin; and the 1.E.19 family is represented by the Clostridium difficile TcdE holin. Toxigenic strains of C. difficile produce two large toxins (TcdA and TcdB) encoded within a pathogenicity locus. tcdE, encoded between tcdA and tcdB, encodes a 166 aa protein which causes death to E. coli when expressed, and the structure of TcdE resembles holins. TcdE acts on the bacterial membrane. Since TcdA and TcdB lack signal peptides, they may be released via TcdE either prior to or subsequent to cell lysis. |
| cd06583 | PGRP | 2.44e-27 | 159 | 289 | 2 | 117 | Peptidoglycan recognition proteins (PGRPs) are pattern recognition receptors that bind, and in certain cases, hydrolyze peptidoglycans (PGNs) of bacterial cell walls. PGRPs have been divided into three classes: short PGRPs (PGRP-S), that are small (20 kDa) extracellular proteins; intermediate PGRPs (PGRP-I) that are 40-45 kDa and are predicted to be transmembrane proteins; and long PGRPs (PGRP-L), up to 90 kDa, which may be either intracellular or transmembrane. Several structures of PGRPs are known in insects and mammals, some bound with substrates like Muramyl Tripeptide (MTP) or Tracheal Cytotoxin (TCT). The substrate binding site is conserved in PGRP-LCx, PGRP-LE, and PGRP-Ialpha proteins. This family includes Zn-dependent N-Acetylmuramoyl-L-alanine Amidase, EC:3.5.1.28. This enzyme cleaves the amide bond between N-acetylmuramoyl and L-amino acids, preferentially D-lactyl-L-Ala, in bacterial cell walls. The structure for the bacteriophage T7 lysozyme shows that two of the conserved histidines and a cysteine are zinc binding residues. Site-directed mutagenesis of T7 lysozyme indicates that two conserved residues, a Tyr and a Lys, are important for amidase activity. |
| COG4824 | COG4824 | 4.78e-27 | 35 | 133 | 30 | 127 | Phage-related holin (Lysis protein) [Mobilome: prophages, transposons]. |
| TIGR01593 | holin_tox_secr | 7.47e-25 | 35 | 133 | 22 | 120 | toxin secretion/phage lysis holin. This model describes one of the many mutally dissimilar families of holins, phage proteins that act together with lytic enzymes in bacterial lysis. This family includes, besides phage holins, the protein TcdE/UtxA involved in toxin secretion in Clostridium difficile and related species. [Protein fate, Protein and peptide secretion and trafficking, Mobile and extrachromosomal element functions, Prophage functions] |
| pfam01510 | Amidase_2 | 9.69e-19 | 158 | 287 | 1 | 112 | N-acetylmuramoyl-L-alanine amidase. This family includes zinc amidases that have N-acetylmuramoyl-L-alanine amidase activity EC:3.5.1.28. This enzyme domain cleaves the amide bond between N-acetylmuramoyl and L-amino acids in bacterial cell walls (preferentially: D-lactyl-L-Ala). The structure is known for the bacteriophage T7 structure and shows that two of the conserved histidines are zinc binding. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AST58407.1 | 1.97e-149 | 138 | 437 | 1 | 293 |
| CUH93073.1 | 5.63e-149 | 138 | 437 | 1 | 293 |
| AUS97328.1 | 2.27e-148 | 138 | 437 | 1 | 293 |
| AEV70121.1 | 3.22e-148 | 138 | 437 | 1 | 293 |
| AEV70155.1 | 6.48e-148 | 138 | 437 | 1 | 293 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3HMB_A | 4.93e-12 | 154 | 285 | 21 | 133 | ChainA, N-acetylmuramoyl-L-alanine amidase xlyA [Bacillus subtilis],3HMB_B Chain B, N-acetylmuramoyl-L-alanine amidase xlyA [Bacillus subtilis],3HMB_C Chain C, N-acetylmuramoyl-L-alanine amidase xlyA [Bacillus subtilis] |
| 3RDR_A | 4.25e-11 | 154 | 285 | 21 | 133 | Structureof the catalytic domain of XlyA [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26835 | 3.94e-23 | 14 | 133 | 12 | 131 | Uncharacterized 14.9 kDa protein in nagH 3'region OS=Clostridium perfringens OX=1502 PE=3 SV=1 |
| P58701 | 3.71e-21 | 8 | 133 | 6 | 131 | Uncharacterized protein CPE0383 OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=CPE0383 PE=3 SV=1 |
| Q92FD1 | 4.24e-21 | 8 | 127 | 3 | 121 | Uncharacterized protein Lin0175 OS=Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) OX=272626 GN=lin0175 PE=3 SV=1 |
| P58702 | 4.24e-21 | 8 | 127 | 3 | 121 | Uncharacterized protein Lmo0128 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=lmo0128 PE=3 SV=1 |
| Q9KE91 | 1.85e-17 | 35 | 132 | 30 | 131 | Uncharacterized protein BH0965 OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=BH0965 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
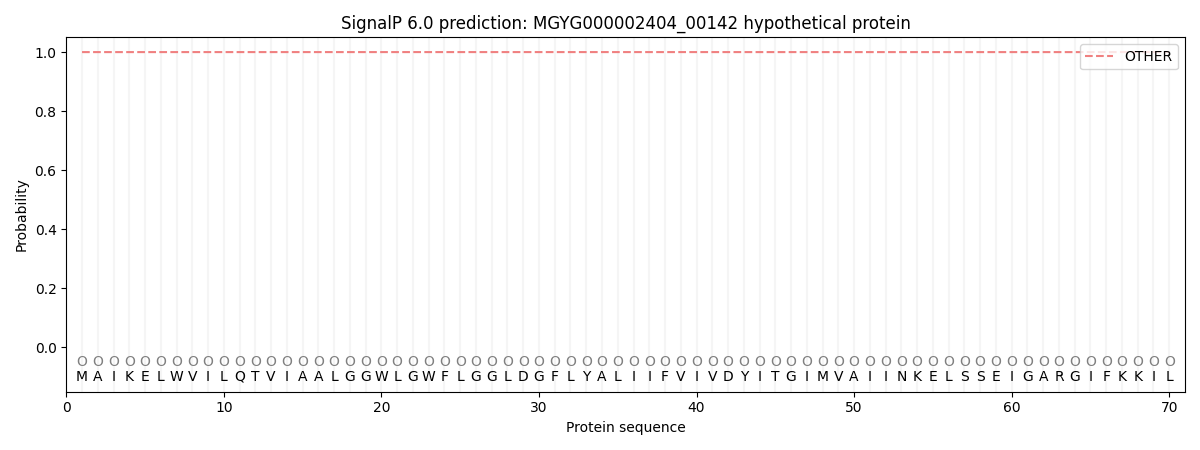
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000030 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |