You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002404_01242
You are here: Home > Sequence: MGYG000002404_01242
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
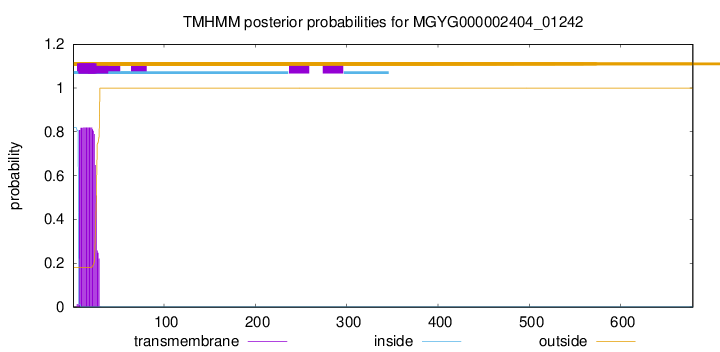
TMHMM annotations
Basic Information help
| Species | Bacillus_BE massilionigeriensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Bacillus_BE; Bacillus_BE massilionigeriensis | |||||||||||
| CAZyme ID | MGYG000002404_01242 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1228023; End: 1230062 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.78e-15 | 329 | 516 | 55 | 236 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 1.81e-11 | 379 | 436 | 1 | 65 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam08239 | SH3_3 | 5.08e-07 | 614 | 675 | 1 | 54 | Bacterial SH3 domain. |
| smart00047 | LYZ2 | 3.02e-06 | 367 | 504 | 1 | 137 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| COG4991 | YraI | 1.03e-05 | 5 | 112 | 1 | 109 | Uncharacterized conserved protein YraI [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSB49442.1 | 1.19e-240 | 1 | 678 | 1 | 701 |
| QIQ32006.1 | 4.78e-240 | 1 | 678 | 1 | 701 |
| AZV41185.1 | 1.15e-235 | 1 | 678 | 1 | 752 |
| QQX25801.1 | 4.30e-232 | 1 | 679 | 1 | 812 |
| ASS87135.1 | 8.91e-228 | 62 | 678 | 1 | 641 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 7.93e-17 | 323 | 501 | 108 | 277 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
| 4PI7_A | 4.87e-09 | 284 | 503 | 4 | 208 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 2.91e-08 | 284 | 503 | 4 | 208 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 3.36e-17 | 236 | 506 | 404 | 650 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 3.62e-17 | 236 | 506 | 448 | 694 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
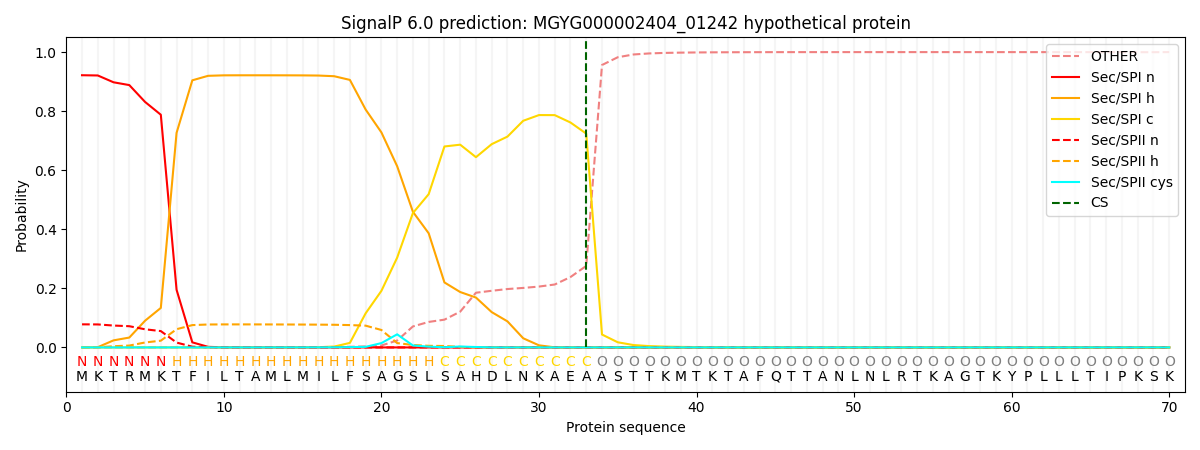
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000657 | 0.919533 | 0.079170 | 0.000243 | 0.000205 | 0.000174 |