You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002406_00086
You are here: Home > Sequence: MGYG000002406_00086
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium pacaense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium pacaense | |||||||||||
| CAZyme ID | MGYG000002406_00086 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43098; End: 44285 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 129 | 349 | 1.7e-42 | 0.9351851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 1.92e-67 | 68 | 383 | 2 | 306 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 1.12e-53 | 68 | 386 | 1 | 315 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 1.34e-41 | 116 | 349 | 46 | 278 | beta-hexosaminidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAC19480.1 | 2.10e-180 | 8 | 389 | 9 | 393 |
| AGG67817.1 | 4.47e-180 | 58 | 391 | 2 | 335 |
| BAV24480.1 | 9.28e-180 | 57 | 390 | 31 | 364 |
| ANR66732.1 | 1.43e-179 | 57 | 390 | 43 | 376 |
| AIK89033.1 | 1.43e-179 | 57 | 390 | 43 | 376 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5IOB_A | 4.49e-174 | 57 | 389 | 16 | 348 | Crystalstructure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_B Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_C Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_D Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_E Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_F Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_G Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032],5IOB_H Crystal structure of beta-N-acetylglucosaminidase-like protein from Corynebacterium glutamicum [Corynebacterium glutamicum ATCC 13032] |
| 4YYF_A | 4.35e-84 | 63 | 392 | 37 | 361 | ChainA, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_B Chain B, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155],4YYF_C Chain C, Beta-N-acetylhexosaminidase [Mycolicibacterium smegmatis MC2 155] |
| 6GFV_A | 8.85e-78 | 63 | 384 | 10 | 333 | Mtuberculosis LpqI [Mycobacterium tuberculosis H37Rv],6GFV_B M tuberculosis LpqI [Mycobacterium tuberculosis H37Rv] |
| 5G1M_A | 2.76e-33 | 95 | 349 | 47 | 293 | Crystalstructure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G1M_B Crystal structure of NagZ from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G2M_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G2M_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with N-acetylglucosamine [Pseudomonas aeruginosa PAO1],5G3R_A Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G3R_B Crystal Structure Of Nagz From Pseudomonas Aeruginosa In Complex With N-acetylglucosamine And L-ala-1,6-anhydromurnac [Pseudomonas aeruginosa PAO1],5G5K_A Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5G5K_B Crystal structure of NagZ from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
| 5G5U_A | 3.74e-32 | 95 | 349 | 47 | 293 | Crystalstructure of NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G5U_B Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G6T_A Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5G6T_B Crystal structure of Zn-containing NagZ H174A mutant from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5LY7_A Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1],5LY7_B Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L7N6B0 | 1.82e-76 | 63 | 384 | 53 | 376 | Beta-hexosaminidase LpqI OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lpqI PE=1 SV=1 |
| A0A0H3M1P5 | 5.10e-76 | 63 | 384 | 53 | 376 | Beta-hexosaminidase LpqI OS=Mycobacterium bovis (strain BCG / Pasteur 1173P2) OX=410289 GN=lpqI PE=3 SV=1 |
| B2FPW9 | 7.87e-34 | 117 | 349 | 45 | 276 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain K279a) OX=522373 GN=nagZ PE=3 SV=1 |
| B4SRK3 | 2.11e-33 | 117 | 349 | 45 | 276 | Beta-hexosaminidase OS=Stenotrophomonas maltophilia (strain R551-3) OX=391008 GN=nagZ PE=3 SV=1 |
| Q9HZK0 | 1.02e-32 | 95 | 349 | 27 | 273 | Beta-hexosaminidase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=nagZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
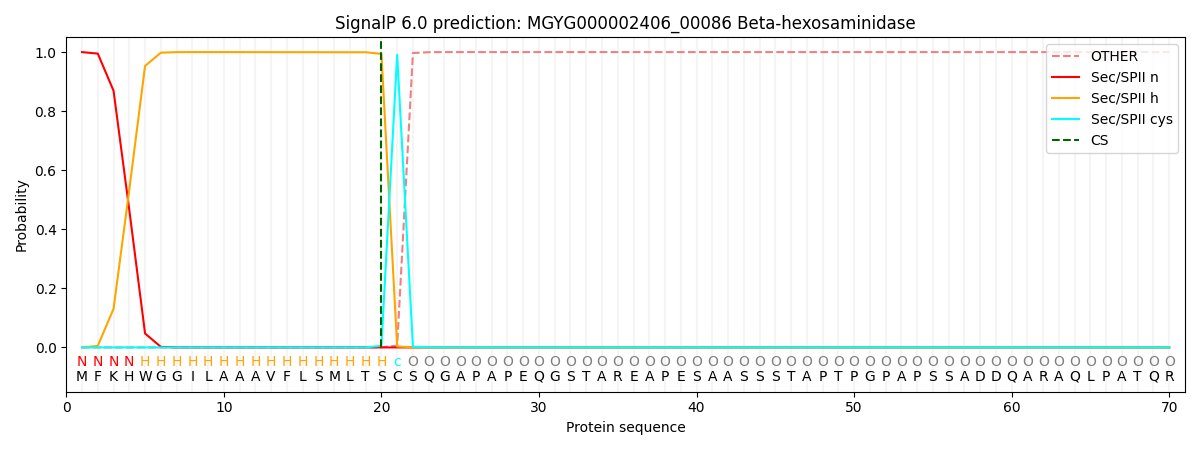
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000022 | 0.000000 | 0.000000 | 0.000000 |
