You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002406_00129
You are here: Home > Sequence: MGYG000002406_00129
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium pacaense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium pacaense | |||||||||||
| CAZyme ID | MGYG000002406_00129 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | Diacylglycerol acyltransferase/mycolyltransferase Ag85C | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 96582; End: 97607 Strand: - | |||||||||||
Full Sequence Download help
| MSVFTRAGAV SRRLIAMLVA LATAAGLMVV GAGTASAANR DWLRPDATGS CDWDPANWWV | 60 |
| QRCDVYSPAM GRNIPVQIQP AQRGGNASLY LLDGMRATDR SNAWLVDTNA AAAYADYNLT | 120 |
| LVMPVGGAGS FYADWNQQAS LSSADPVIYM WETFLTSELP AYLEANFGVA RNNNSIAGLS | 180 |
| MGGTAALNLA AKHPAQFRQA MSWSGYLNTT APGMQTMLRL AMLDTGGFNV NAMYGSIINP | 240 |
| RRFENDPFWN MAGLHGTDVY ISAASGLWGP ADNGTRVDHR MTGSVLEFVA MTSTRAWEAK | 300 |
| ARLEGLNPTA DYPVTGIHSW AQFNSQLYNT RERVLNVMNA W | 341 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 67 | 330 | 7.5e-44 | 0.960352422907489 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0627 | FrmB | 3.01e-65 | 12 | 341 | 4 | 315 | S-formylglutathione hydrolase FrmB [Defense mechanisms]. |
| pfam00756 | Esterase | 6.92e-30 | 67 | 327 | 1 | 239 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| COG2382 | Fes | 0.001 | 154 | 215 | 157 | 220 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMA01272.1 | 1.97e-216 | 1 | 341 | 1 | 341 |
| BAU97240.1 | 1.14e-215 | 1 | 341 | 1 | 341 |
| BAC19520.1 | 6.57e-215 | 1 | 341 | 20 | 360 |
| ANE05042.1 | 6.57e-215 | 1 | 341 | 1 | 341 |
| ALC06933.1 | 9.33e-215 | 1 | 341 | 1 | 341 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MQL_A | 1.12e-55 | 60 | 340 | 10 | 284 | ChainA, Diacylglycerol acyltransferase/mycolyltransferase Ag85C [Mycobacterium tuberculosis] |
| 7MYG_A | 2.13e-55 | 60 | 340 | 4 | 278 | ChainA, Diacylglycerol acyltransferase [Mycobacterium tuberculosis],7MYG_B Chain B, Diacylglycerol acyltransferase [Mycobacterium tuberculosis] |
| 1DQZ_A | 2.26e-55 | 60 | 340 | 6 | 280 | ChainA, PROTEIN (ANTIGEN 85-C) [Mycobacterium tuberculosis],1DQZ_B Chain B, PROTEIN (ANTIGEN 85-C) [Mycobacterium tuberculosis] |
| 1DQY_A | 2.46e-55 | 60 | 340 | 9 | 283 | ChainA, PROTEIN (ANTIGEN 85-C) [Mycobacterium tuberculosis] |
| 4QDX_A | 4.08e-55 | 60 | 340 | 9 | 283 | Crystalstructure of Antigen 85C-C209G mutant [Mycobacterium tuberculosis H37Rv] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WQN9 | 8.99e-54 | 60 | 340 | 54 | 328 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=fbpC PE=1 SV=1 |
| P0A4V5 | 8.99e-54 | 60 | 340 | 54 | 328 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=fbpC PE=3 SV=1 |
| P9WQN8 | 8.99e-54 | 60 | 340 | 54 | 328 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=fbpC PE=3 SV=1 |
| Q05862 | 4.89e-51 | 60 | 340 | 54 | 328 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium leprae (strain TN) OX=272631 GN=fbpC PE=3 SV=1 |
| O52972 | 1.11e-50 | 60 | 327 | 54 | 315 | Diacylglycerol acyltransferase/mycolyltransferase Ag85C OS=Mycobacterium avium OX=1764 GN=fbpC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
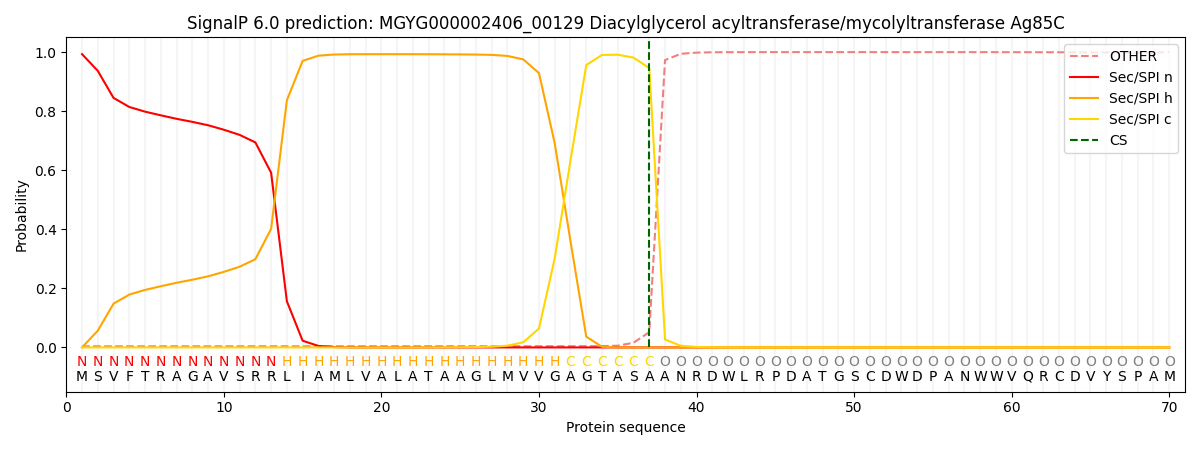
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006526 | 0.988175 | 0.000372 | 0.004232 | 0.000398 | 0.000266 |

