You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002406_00547
You are here: Home > Sequence: MGYG000002406_00547
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium pacaense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium pacaense | |||||||||||
| CAZyme ID | MGYG000002406_00547 | |||||||||||
| CAZy Family | GT85 | |||||||||||
| CAZyme Description | Galactan 5-O-arabinofuranosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 579342; End: 581402 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT85 | 49 | 496 | 1.8e-153 | 0.990632318501171 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12250 | AftA_N | 0.0 | 48 | 493 | 1 | 424 | Arabinofuranosyltransferase N terminal. This domain family is found in bacteria, and is typically between 430 and 441 amino acids in length. This family is the N terminal region of AftA. The enzyme catalyzes the addition of the first key arabinofuranosyl residue from the sugar donor beta-D-arabinofuranosyl-1-monophosphoryldecaprenol to the galactan domain of the cell wall, thus priming the galactan for further elaboration by the arabinofuranosyltransferases. The N terminal region has been predicted to span 11 transmembrane regions. |
| pfam12249 | AftA_C | 8.09e-101 | 503 | 684 | 1 | 177 | Arabinofuranosyltransferase A C terminal. This domain family is found in bacteria, and is typically between 179 and 190 amino acids in length. This family is the C terminal region of AftA. The enzyme catalyzes the addition of the first key arabinofuranosyl residue from the sugar donor beta-D-arabinofuranosyl-1-monophosphoryldecaprenol to the galactan domain of the cell wall, thus priming the galactan for further elaboration by the arabinofuranosyltransferases. The C terminal region is predicted to be directed towards the periplasm. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAC16972.1 | 0.0 | 1 | 686 | 1 | 703 |
| AGG65634.1 | 8.97e-317 | 1 | 686 | 1 | 677 |
| ALC04697.1 | 2.16e-316 | 1 | 686 | 1 | 681 |
| BAB97581.1 | 3.92e-314 | 35 | 686 | 15 | 661 |
| ASW12977.1 | 7.03e-314 | 35 | 686 | 31 | 677 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8NTW4 | 7.84e-315 | 35 | 686 | 15 | 661 | Galactan 5-O-arabinofuranosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=aftA PE=1 SV=1 |
| A0R611 | 2.75e-117 | 141 | 684 | 80 | 613 | Galactan 5-O-arabinofuranosyltransferase OS=Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) OX=246196 GN=aftA PE=3 SV=1 |
| P9WN02 | 2.69e-107 | 46 | 684 | 32 | 637 | Galactan 5-O-arabinofuranosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=aftA PE=3 SV=1 |
| P9WN03 | 2.69e-107 | 46 | 684 | 32 | 637 | Galactan 5-O-arabinofuranosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=aftA PE=1 SV=1 |
| Q7TVN5 | 2.69e-107 | 46 | 684 | 32 | 637 | Galactan 5-O-arabinofuranosyltransferase OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=aftA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
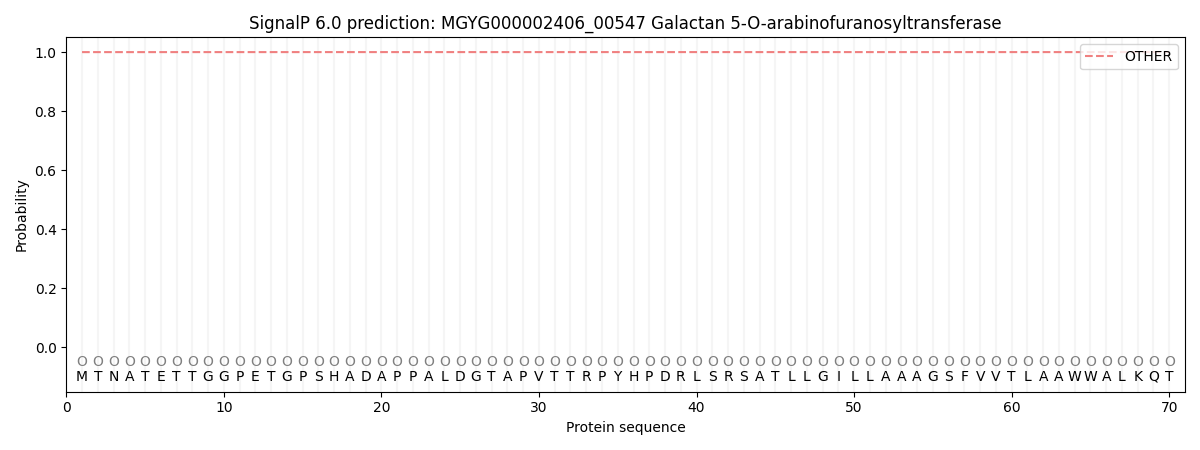
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000005 | 0.000032 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
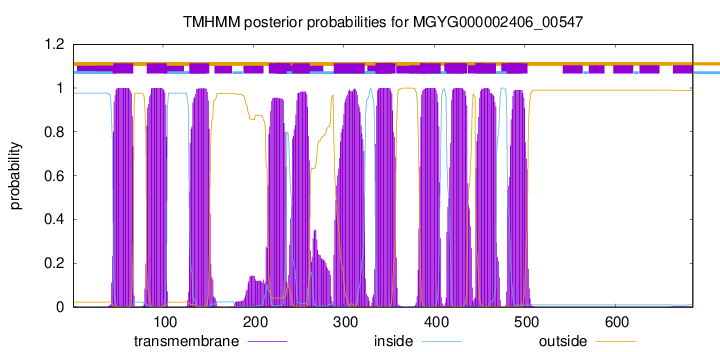
| start | end |
|---|---|
| 45 | 67 |
| 82 | 104 |
| 129 | 151 |
| 217 | 236 |
| 243 | 262 |
| 289 | 323 |
| 335 | 357 |
| 385 | 407 |
| 414 | 436 |
| 446 | 468 |
| 481 | 503 |
