You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002407_02538
You are here: Home > Sequence: MGYG000002407_02538
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cohnella sp900169535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Cohnella; Cohnella sp900169535 | |||||||||||
| CAZyme ID | MGYG000002407_02538 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 706795; End: 713403 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14953 | NHL_like_1 | 4.22e-119 | 148 | 533 | 1 | 323 | Uncharacterized NHL-repeat domain in bacterial proteins. This bacterial family of NHL-repeat domains is found in a variety of domain architectures. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
| cd14953 | NHL_like_1 | 5.53e-118 | 93 | 478 | 2 | 323 | Uncharacterized NHL-repeat domain in bacterial proteins. This bacterial family of NHL-repeat domains is found in a variety of domain architectures. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
| cd14953 | NHL_like_1 | 8.61e-116 | 317 | 700 | 1 | 323 | Uncharacterized NHL-repeat domain in bacterial proteins. This bacterial family of NHL-repeat domains is found in a variety of domain architectures. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
| cd14953 | NHL_like_1 | 9.75e-104 | 41 | 367 | 1 | 323 | Uncharacterized NHL-repeat domain in bacterial proteins. This bacterial family of NHL-repeat domains is found in a variety of domain architectures. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
| cd14953 | NHL_like_1 | 9.30e-67 | 36 | 310 | 104 | 323 | Uncharacterized NHL-repeat domain in bacterial proteins. This bacterial family of NHL-repeat domains is found in a variety of domain architectures. The NHL (NCL-1, HT2A and LIN-41) repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBH18999.1 | 2.75e-111 | 1786 | 2202 | 1220 | 1638 |
| QNK56141.1 | 1.13e-106 | 1787 | 2202 | 1532 | 1949 |
| ANY68648.1 | 3.78e-102 | 1786 | 2202 | 1494 | 1912 |
| QCT04045.1 | 1.20e-99 | 1776 | 2202 | 1181 | 1611 |
| ANY68649.1 | 2.61e-97 | 1787 | 2202 | 1242 | 1661 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SKE_A | 3.05e-19 | 219 | 533 | 211 | 552 | Teneurin2 in complex with Latrophilin 2 Lec domain [Gallus gallus],6SKE_C Teneurin 2 in complex with Latrophilin 2 Lec domain [Gallus gallus] |
| 6SKA_A | 3.10e-19 | 219 | 533 | 295 | 636 | Teneurin2 in complex with Latrophilin 1 Lec-Olf domains [Gallus gallus] |
| 6FB3_A | 3.11e-19 | 219 | 533 | 305 | 646 | Teneurin2 Partial Extracellular Domain [Gallus gallus],6FB3_B Teneurin 2 Partial Extracellular Domain [Gallus gallus],6FB3_C Teneurin 2 Partial Extracellular Domain [Gallus gallus],6FB3_D Teneurin 2 Partial Extracellular Domain [Gallus gallus] |
| 6VHH_A | 1.78e-17 | 219 | 533 | 384 | 718 | HumanTeneurin-2 and human Latrophilin-3 binary complex [Homo sapiens] |
| 6CMX_A | 1.80e-17 | 219 | 533 | 461 | 795 | HumanTeneurin 2 extra-cellular region [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 6.38e-27 | 1813 | 2199 | 1501 | 1858 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| C6CRV0 | 3.24e-25 | 1979 | 2197 | 1244 | 1460 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P38535 | 1.01e-24 | 2003 | 2200 | 894 | 1085 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q8VZ10 | 4.46e-19 | 111 | 423 | 562 | 885 | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic OS=Arabidopsis thaliana OX=3702 GN=SOQ1 PE=1 SV=1 |
| Q9DER5 | 1.87e-18 | 219 | 533 | 1257 | 1598 | Teneurin-2 OS=Gallus gallus OX=9031 GN=TENM2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
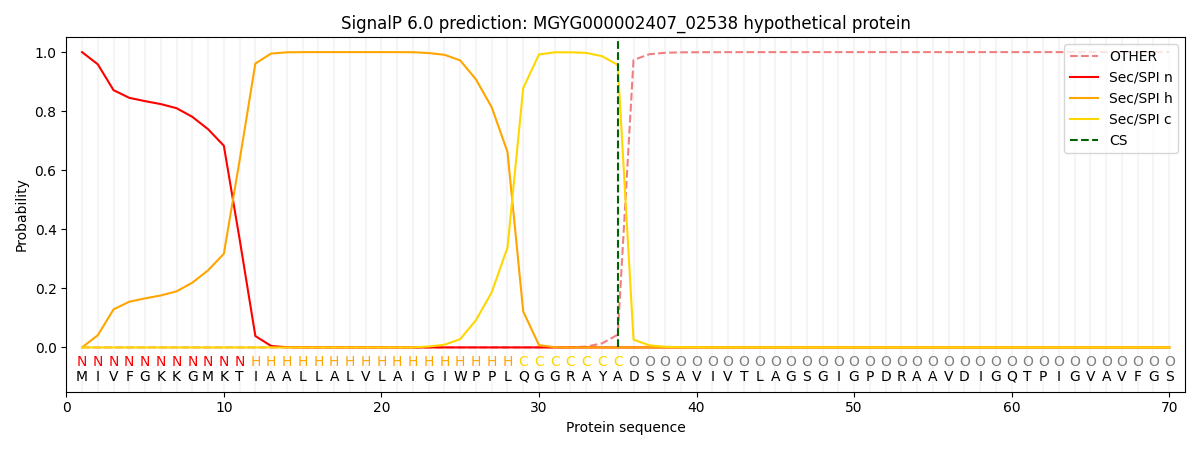
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000381 | 0.998828 | 0.000207 | 0.000197 | 0.000176 | 0.000159 |

