You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002407_04446
You are here: Home > Sequence: MGYG000002407_04446
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cohnella sp900169535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Cohnella; Cohnella sp900169535 | |||||||||||
| CAZyme ID | MGYG000002407_04446 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1855816; End: 1857768 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1835 | OafA | 8.30e-64 | 3 | 357 | 6 | 358 | Peptidoglycan/LPS O-acetylase OafA/YrhL, contains acyltransferase and SGNH-hydrolase domains [Cell wall/membrane/envelope biogenesis]. |
| cd01840 | SGNH_hydrolase_yrhL_like | 1.46e-59 | 499 | 648 | 1 | 150 | yrhL-like subfamily of SGNH-hydrolases, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. Most members of this sub-family appear to co-occur with N-terminal acyltransferase domains. Might be involved in lipid metabolism. |
| pfam01757 | Acyl_transf_3 | 7.08e-22 | 12 | 349 | 1 | 326 | Acyltransferase family. This family includes a range of acyltransferase enzymes. This domain is found in many as yet uncharacterized C. elegans proteins and it is approximately 300 amino acids long. |
| cd00229 | SGNH_hydrolase | 4.72e-07 | 500 | 644 | 1 | 184 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| pfam13472 | Lipase_GDSL_2 | 0.007 | 552 | 639 | 65 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQO07014.1 | 1.33e-114 | 9 | 640 | 27 | 632 |
| QNQ36362.1 | 2.65e-54 | 2 | 404 | 50 | 440 |
| ARI54412.1 | 8.99e-53 | 4 | 447 | 33 | 463 |
| QTI42935.1 | 9.20e-53 | 4 | 404 | 35 | 425 |
| BAG21593.1 | 1.01e-52 | 2 | 404 | 55 | 445 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6VJP_A | 2.36e-31 | 502 | 650 | 7 | 155 | ChainA, Acetyltransferase [Staphylococcus aureus],6VJP_B Chain B, Acetyltransferase [Staphylococcus aureus] |
| 6WN9_A | 2.12e-30 | 502 | 650 | 7 | 155 | ChainA, Acetyltransferase [Staphylococcus aureus],6WN9_B Chain B, Acetyltransferase [Staphylococcus aureus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O05402 | 5.67e-143 | 7 | 650 | 6 | 634 | Putative peptidoglycan O-acetyltransferase YrhL OS=Bacillus subtilis (strain 168) OX=224308 GN=yrhL PE=3 SV=1 |
| Q8Y7I6 | 4.83e-138 | 1 | 645 | 1 | 617 | Peptidoglycan O-acetyltransferase OatA OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=oatA PE=1 SV=1 |
| Q6GDN2 | 7.81e-106 | 10 | 650 | 16 | 597 | O-acetyltransferase OatA OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=oatA PE=3 SV=1 |
| Q7A3D6 | 2.17e-105 | 10 | 650 | 16 | 597 | O-acetyltransferase OatA OS=Staphylococcus aureus (strain N315) OX=158879 GN=oatA PE=1 SV=1 |
| Q79ZY2 | 2.17e-105 | 10 | 650 | 16 | 597 | O-acetyltransferase OatA OS=Staphylococcus aureus (strain MW2) OX=196620 GN=oatA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
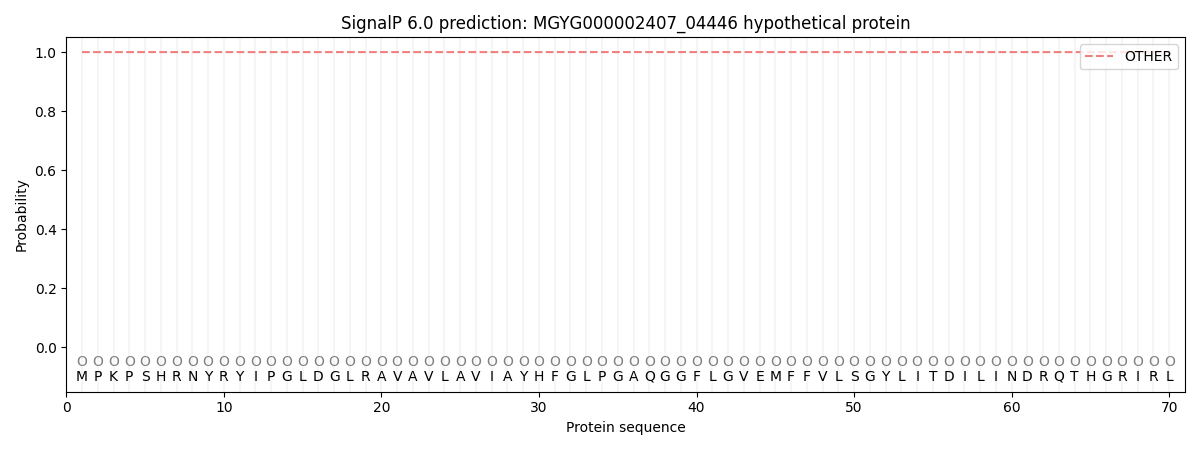
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000033 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
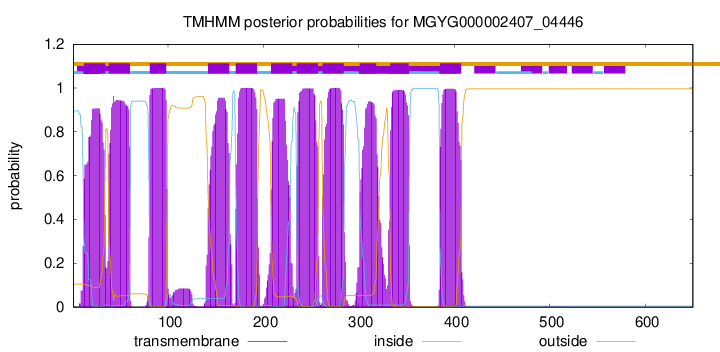
| start | end |
|---|---|
| 12 | 34 |
| 38 | 60 |
| 81 | 98 |
| 142 | 164 |
| 171 | 193 |
| 208 | 230 |
| 235 | 257 |
| 262 | 284 |
| 301 | 318 |
| 333 | 352 |
| 385 | 407 |
