You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002408_02691
You are here: Home > Sequence: MGYG000002408_02691
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus_BD tuaregi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-18226; Bacillus_BD; Bacillus_BD tuaregi | |||||||||||
| CAZyme ID | MGYG000002408_02691 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2259941; End: 2260552 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3773 | CwlJ | 7.04e-43 | 78 | 203 | 110 | 242 | Cell wall hydrolase CwlJ, involved in spore germination [Cell cycle control, cell division, chromosome partitioning, Cell wall/membrane/envelope biogenesis]. |
| pfam07486 | Hydrolase_2 | 1.62e-42 | 100 | 202 | 1 | 101 | Cell Wall Hydrolase. These enzymes have been implicated in cell wall hydrolysis, most extensively in Bacillus subtilis. For instance Bacillus subtilis steB is expressed during sporulation as an inactive form and then deposited on the cell outer cortex. During germination the the enzyme is activated and hydrolyzes the cortex. A similar role is carried out by the partially redundant Bacillus subtilis CwlJ. It is not clear whether these enzymes are amidases or peptidases. |
| pfam01476 | LysM | 2.69e-14 | 36 | 78 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| smart00257 | LysM | 3.30e-13 | 35 | 77 | 1 | 44 | Lysin motif. |
| TIGR02899 | spore_safA | 5.18e-13 | 38 | 78 | 1 | 43 | spore coat assembly protein SafA. SafA (YrbB) (SafA) of Bacillus subtilis is a protein found at the interface of the spore cortex and spore coat, and is dependent on SpoVID for its localization. This model is based on the N-terminal LysM (lysin motif) domain (see pfamAM model pfam01476) of SafA and, from several other spore-forming species, the protein with the most similar N-terminal region. However, this set of proteins differs greatly in C-terminal of the LysM domaim; blocks of 12-residue and 13-residue repeats are found in the Bacillus cereus group, tandem LysM domains in Thermoanaerobacter tengcongensis MB4, etc. in which one of which is found in most examples of endospore-forming bacteria. [Cellular processes, Sporulation and germination] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCJ43879.1 | 1.65e-84 | 1 | 203 | 1 | 203 |
| AYA75830.1 | 2.29e-83 | 1 | 203 | 1 | 198 |
| AGK55415.1 | 1.29e-80 | 1 | 203 | 1 | 199 |
| QNU33833.1 | 6.96e-80 | 1 | 203 | 1 | 197 |
| QSB48397.1 | 6.96e-80 | 1 | 203 | 1 | 197 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4F55_A | 6.25e-25 | 83 | 203 | 12 | 128 | CrystalStructure of the Catalytic Domain of the Bacillus cereus SleB Protein [Bacillus cereus ATCC 14579] |
| 4FET_A | 6.79e-23 | 83 | 203 | 106 | 222 | Catalyticdomain of germination-specific lytic tansglycosylase SleB from Bacillus anthracis [Bacillus anthracis],4FET_B Catalytic domain of germination-specific lytic tansglycosylase SleB from Bacillus anthracis [Bacillus anthracis] |
| 2MKX_A | 1.21e-06 | 36 | 81 | 7 | 52 | Solutionstructure of LysM the peptidoglycan binding domain of autolysin AtlA from Enterococcus faecalis [Enterococcus faecalis V583] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31685 | 2.03e-33 | 79 | 202 | 88 | 207 | Uncharacterized protein YkvT OS=Bacillus subtilis (strain 168) OX=224308 GN=ykvT PE=2 SV=1 |
| Q9KCE0 | 1.58e-25 | 83 | 201 | 214 | 328 | Spore cortex-lytic enzyme OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=sleB PE=3 SV=1 |
| P0A3V0 | 7.33e-23 | 83 | 203 | 143 | 259 | Spore cortex-lytic enzyme OS=Bacillus cereus (strain ATCC 14579 / DSM 31 / CCUG 7414 / JCM 2152 / NBRC 15305 / NCIMB 9373 / NCTC 2599 / NRRL B-3711) OX=226900 GN=sleB PE=1 SV=1 |
| P0A3V1 | 7.33e-23 | 83 | 203 | 143 | 259 | Spore cortex-lytic enzyme OS=Bacillus cereus OX=1396 GN=sleB PE=1 SV=1 |
| P50739 | 5.97e-22 | 83 | 201 | 189 | 303 | Spore cortex-lytic enzyme OS=Bacillus subtilis (strain 168) OX=224308 GN=sleB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
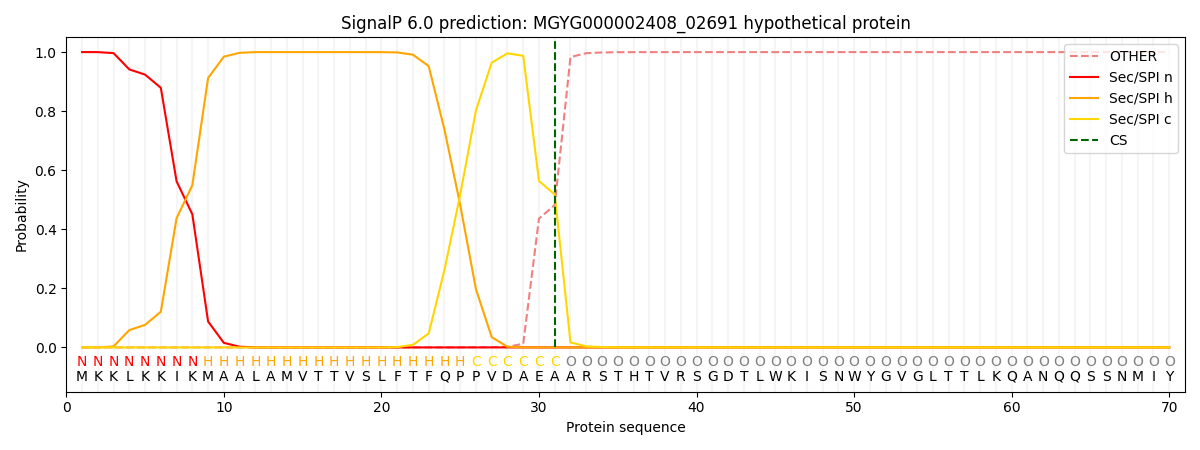
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000268 | 0.999009 | 0.000195 | 0.000186 | 0.000166 | 0.000148 |
