You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_00653
You are here: Home > Sequence: MGYG000002414_00653
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_00653 | |||||||||||
| CAZy Family | CBM16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 733430; End: 735685 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM16 | 97 | 220 | 5.9e-18 | 0.9224137931034483 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5542 | COG5542 | 1.05e-28 | 283 | 655 | 29 | 369 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| pfam09594 | GT87 | 5.64e-08 | 374 | 623 | 3 | 235 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
| COG1807 | ArnT | 1.27e-07 | 363 | 643 | 54 | 330 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 2.45e-05 | 373 | 533 | 1 | 159 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| pfam02018 | CBM_4_9 | 3.06e-04 | 96 | 215 | 1 | 101 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIQ72361.1 | 0.0 | 1 | 751 | 1 | 751 |
| AIQ33730.1 | 0.0 | 68 | 751 | 1 | 684 |
| AIQ21956.1 | 0.0 | 68 | 747 | 1 | 680 |
| CQR52353.1 | 0.0 | 68 | 743 | 1 | 677 |
| QSF45552.1 | 0.0 | 68 | 740 | 1 | 673 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000091 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
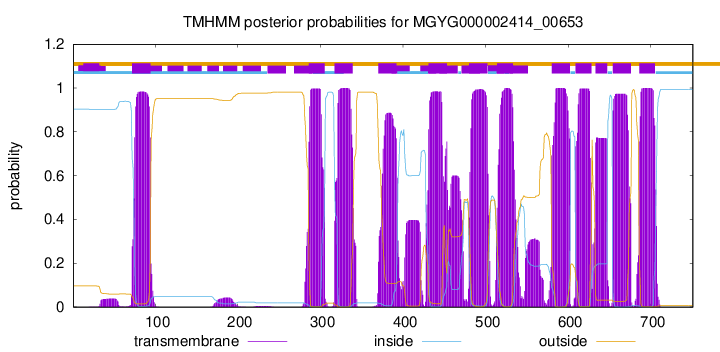
| start | end |
|---|---|
| 72 | 94 |
| 286 | 305 |
| 317 | 339 |
| 370 | 392 |
| 431 | 453 |
| 480 | 502 |
| 514 | 533 |
| 580 | 602 |
| 609 | 628 |
| 633 | 647 |
| 654 | 676 |
| 686 | 705 |
