You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_00963
You are here: Home > Sequence: MGYG000002414_00963
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
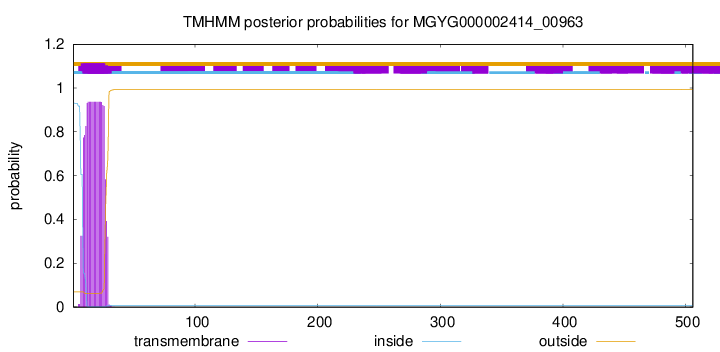
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_00963 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1110451; End: 1111971 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 48 | 346 | 3.4e-84 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 3.33e-49 | 49 | 348 | 2 | 311 | Glycosyl hydrolase family 26. |
| smart00758 | PA14 | 2.72e-35 | 367 | 500 | 1 | 135 | domain in bacterial beta-glucosidases other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins. |
| pfam07691 | PA14 | 1.43e-34 | 367 | 502 | 3 | 141 | PA14 domain. This domain forms an insert in bacterial beta-glucosidases and is found in other glycosidases, glycosyltransferases, proteases, amidases, yeast adhesins, and bacterial toxins, including anthrax protective antigen (PA). The domain also occurs in a Dictyostelium prespore-cell-inducing factor Psi and in fibrocystin, the mammalian protein whose mutation leads to polycystic kidney and hepatic disease. The crystal structure of PA shows that this domain (named PA14 after its location in the PA20 pro-peptide) has a beta-barrel structure. The PA14 domain sequence suggests a binding function, rather than a catalytic role. The PA14 domain distribution is compatible with carbohydrate binding. |
| COG4124 | ManB2 | 1.02e-18 | 174 | 300 | 160 | 296 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIQ72622.1 | 0.0 | 1 | 506 | 1 | 506 |
| AWV36570.1 | 0.0 | 14 | 506 | 1 | 493 |
| AIQ22237.1 | 0.0 | 1 | 506 | 1 | 506 |
| AIQ33983.1 | 0.0 | 1 | 506 | 1 | 503 |
| AIQ27762.1 | 2.56e-265 | 5 | 506 | 6 | 506 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q75_A | 1.95e-74 | 47 | 352 | 23 | 330 | Thestructure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75],6Q75_B The structure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75] |
| 3ZM8_A | 1.02e-42 | 49 | 352 | 147 | 443 | ChainA, Gh26 Endo-beta-1,4-mannanase [Podospora anserina S mat+] |
| 6HPF_A | 4.50e-41 | 43 | 348 | 2 | 309 | Structureof Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose [Yunnania penicillata] |
| 6QE7_A | 7.14e-40 | 368 | 504 | 5 | 140 | ChainA, Anti-sigma-I factor RsgI3 [Acetivibrio thermocellus],6QE7_B Chain B, Anti-sigma-I factor RsgI3 [Acetivibrio thermocellus],6QE7_C Chain C, Anti-sigma-I factor RsgI3 [Acetivibrio thermocellus] |
| 3WDQ_A | 8.13e-38 | 47 | 348 | 34 | 351 | Crystalstructure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus [Symbiotic protist of Reticulitermes speratus],3WDR_A Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus complexed with gluco-manno-oligosaccharide [Symbiotic protist of Reticulitermes speratus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2Q4H7 | 1.39e-45 | 49 | 352 | 176 | 472 | Mannan endo-1,4-beta-mannosidase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Man26A PE=1 SV=1 |
| Q5AWB7 | 1.25e-38 | 23 | 322 | 7 | 320 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
| A3DC75 | 7.68e-37 | 367 | 504 | 357 | 493 | Anti-sigma-I factor RsgI3 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI3 PE=1 SV=1 |
| P55296 | 3.59e-36 | 35 | 348 | 146 | 459 | Mannan endo-1,4-beta-mannosidase A OS=Piromyces sp. OX=45796 GN=MANA PE=2 SV=1 |
| P55297 | 5.74e-35 | 35 | 322 | 147 | 431 | Mannan endo-1,4-beta-mannosidase B OS=Piromyces sp. OX=45796 GN=MANB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
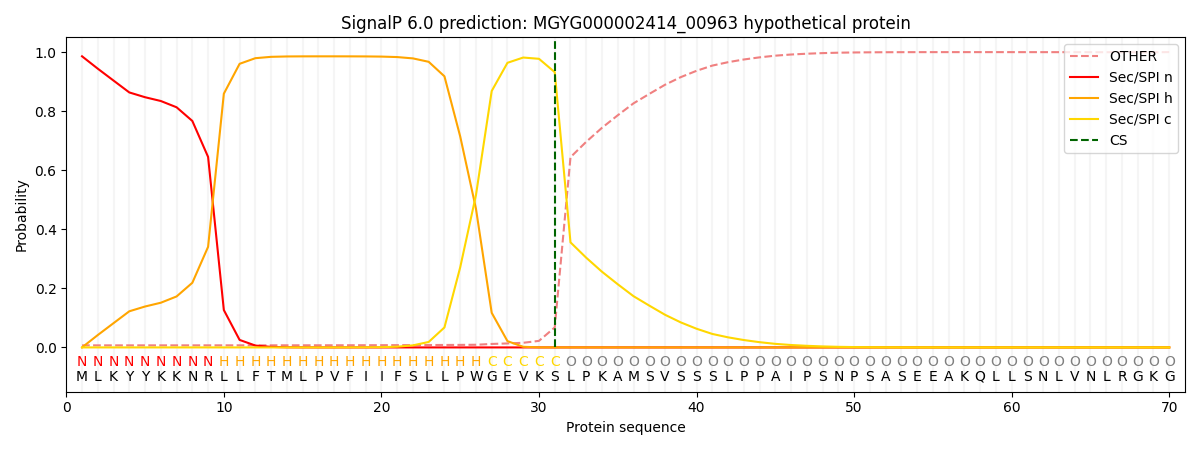
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009949 | 0.980900 | 0.008230 | 0.000383 | 0.000255 | 0.000263 |