You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_00964
You are here: Home > Sequence: MGYG000002414_00964
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
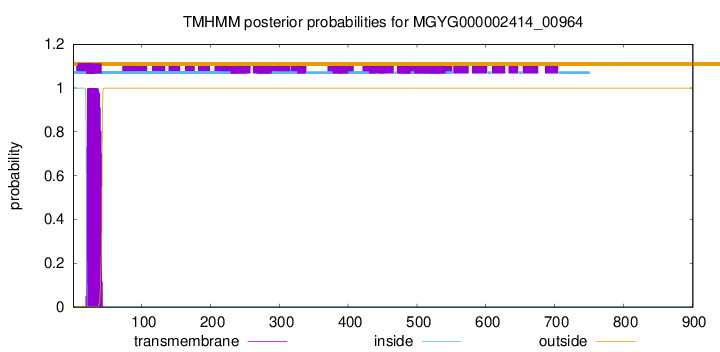
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_00964 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1112023; End: 1114728 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 780 | 896 | 1e-22 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.80e-16 | 780 | 895 | 4 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| smart00060 | FN3 | 2.12e-04 | 683 | 752 | 1 | 76 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| cd00063 | FN3 | 2.68e-04 | 683 | 751 | 1 | 75 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| pfam14683 | CBM-like | 0.005 | 72 | 228 | 1 | 157 | Polysaccharide lyase family 4, domain III. CBM-like is domain III of rhamnogalacturonan lyase (RG-lyase). The full-length protein specifically recognizes and cleaves alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in the backbone of rhamnogalacturonan-I, a major component of the plant cell wall polysaccharide, pectin. This domain possesses a jelly roll beta-sandwich fold structurally homologous to carbohydrate binding modules (CBMs), and it carries two sulfate ions and a hexa-coordinated calcium ion. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV31987.1 | 0.0 | 1 | 901 | 1 | 901 |
| AIQ72623.1 | 0.0 | 54 | 901 | 1 | 848 |
| AIQ22238.1 | 0.0 | 54 | 901 | 1 | 848 |
| AIQ33984.1 | 0.0 | 54 | 901 | 1 | 848 |
| AIQ50890.1 | 0.0 | 6 | 901 | 2 | 896 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5G56_A | 1.14e-06 | 668 | 821 | 604 | 755 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 3MPC_A | 1.83e-06 | 683 | 750 | 5 | 74 | ChainA, Fn3-like protein [Acetivibrio thermocellus ATCC 27405],3MPC_B Chain B, Fn3-like protein [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
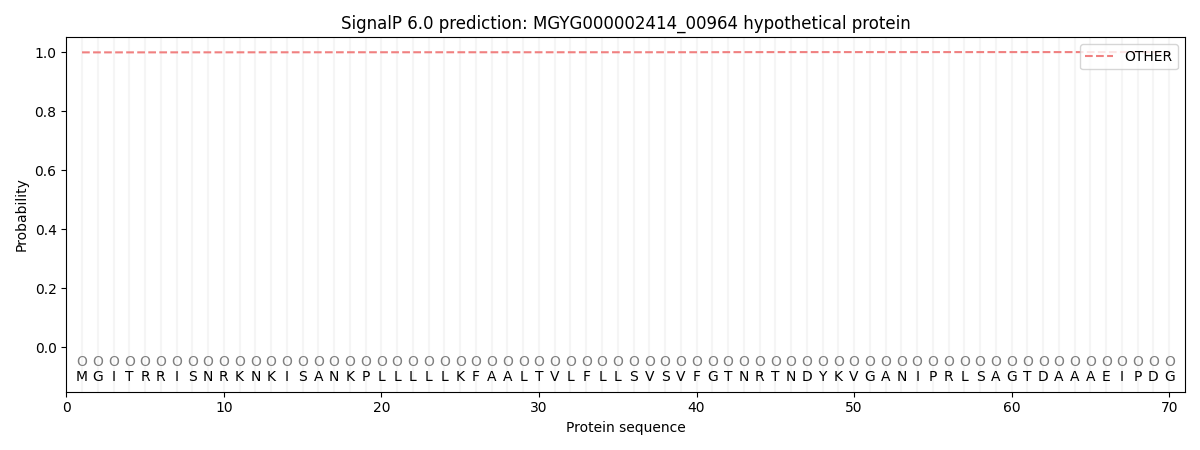
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999227 | 0.000702 | 0.000007 | 0.000003 | 0.000002 | 0.000077 |