You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_01362
You are here: Home > Sequence: MGYG000002414_01362
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
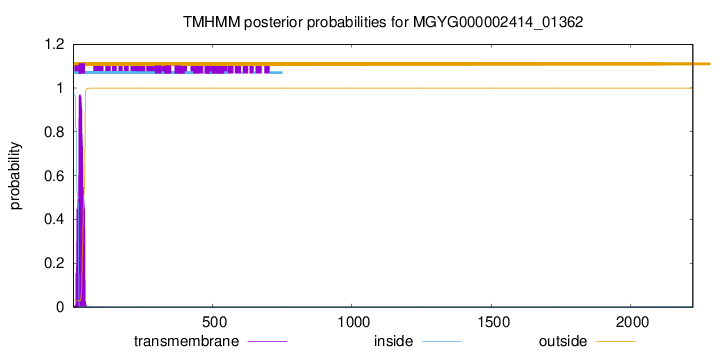
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_01362 | |||||||||||
| CAZy Family | GH59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1541872; End: 1548546 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH59 | 326 | 1055 | 8.7e-192 | 0.993660855784469 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02057 | Glyco_hydro_59 | 1.48e-101 | 332 | 658 | 1 | 292 | Glycosyl hydrolase family 59. |
| pfam16403 | DUF5011 | 9.59e-18 | 1627 | 1705 | 1 | 69 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
| pfam16403 | DUF5011 | 1.92e-17 | 1718 | 1798 | 1 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
| NF033190 | inl_like_NEAT_1 | 3.08e-17 | 2047 | 2219 | 582 | 750 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 1.83e-09 | 2106 | 2147 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUL53185.1 | 0.0 | 1 | 2223 | 1 | 2224 |
| QNK59157.1 | 0.0 | 2 | 1522 | 3 | 1539 |
| QOR70567.1 | 0.0 | 18 | 1593 | 40 | 1629 |
| ACL75594.1 | 0.0 | 306 | 1191 | 26 | 910 |
| ABG76970.1 | 0.0 | 306 | 1191 | 26 | 910 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 3.82e-20 | 2045 | 2218 | 24 | 193 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 3.92e-20 | 2045 | 2218 | 3 | 172 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 3.78e-42 | 1871 | 2221 | 1502 | 1855 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 6.59e-30 | 2040 | 2221 | 901 | 1081 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P19424 | 5.75e-26 | 2047 | 2221 | 41 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| C6CRV0 | 4.82e-22 | 1932 | 2217 | 1172 | 1455 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P38537 | 1.63e-17 | 2055 | 2221 | 42 | 206 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
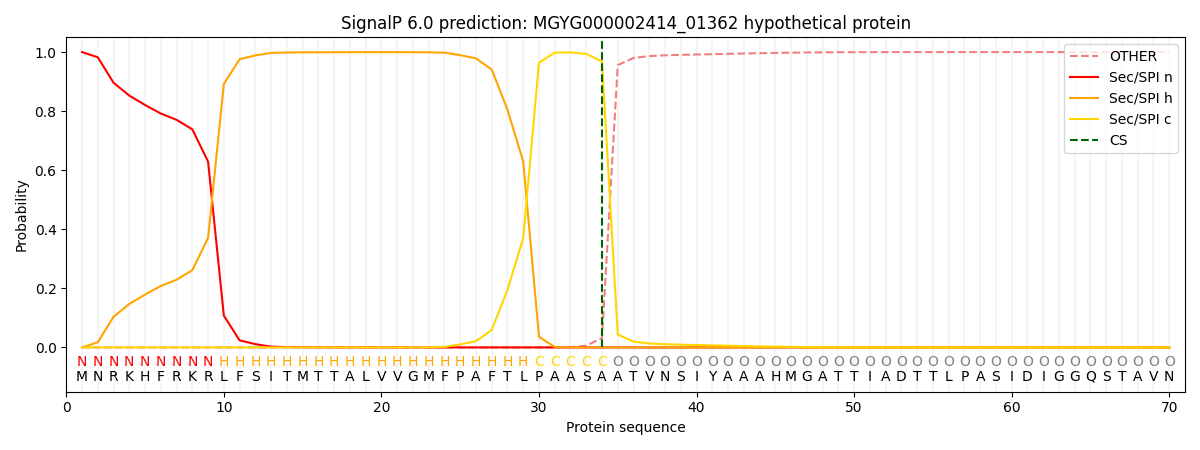
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000535 | 0.998571 | 0.000208 | 0.000282 | 0.000197 | 0.000167 |