You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_01900
You are here: Home > Sequence: MGYG000002414_01900
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_01900 | |||||||||||
| CAZy Family | GH68 | |||||||||||
| CAZyme Description | Levansucrase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2195500; End: 2196984 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH68 | 43 | 486 | 1e-130 | 0.9928057553956835 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02435 | Glyco_hydro_68 | 6.15e-144 | 43 | 476 | 1 | 409 | Levansucrase/Invertase. This Pfam family consists of the glycosyl hydrolase 68 family, including several bacterial levansucrase enzymes, and invertase from zymomonas. |
| cd08997 | GH68 | 2.05e-117 | 95 | 476 | 1 | 354 | Glycosyl hydrolase family 68, includes levansucrase, beta-fructofuranosidase and inulosucrase. Glycosyl hydrolase family 68 (GH68) consists of frucosyltransferases (FTFs) that include levansucrase (EC 2.4.1.10), beta-fructofuranosidase (EC 3.2.1.26) and inulosucrase (EC 2.4.1.9), all of which use sucrose as their preferential donor substrate. Levansucrase, also known as beta-D-fructofuranosyl transferase, catalyzes the transfer of the sucrose fructosyl moiety to a growing levan chain. Similarly, inulosucrase catalyzes long inulin-type of fructans, and beta-fructofuranosidases create fructooligosaccharides (FOS). However, in the absence of high fructan/sucrose ratio, some GH68 enzymes can also use fructan as donor substrate. GH68 retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. Biotechnological applications of these enzymes include use of inulin in inexpensive production of rich fructose syrups as well as use of FOS as health-promoting pre-biotics. |
| cd08979 | GH_J | 1.80e-62 | 96 | 467 | 1 | 292 | Glycosyl hydrolase families 32 and 68, which form the clan GH-J. This glycosyl hydrolase family clan J (according to carbohydrate-active enzymes database (CAZY)) includes family 32 (GH32) and 68 (GH68). GH32 enzymes include invertase (EC 3.2.1.26) and other other fructofuranosidases such as inulinase (EC 3.2.1.7), exo-inulinase (EC 3.2.1.80), levanase (EC 3.2.1.65), and transfructosidases such sucrose:sucrose 1-fructosyltransferase (EC 2.4.1.99), fructan:fructan 1-fructosyltransferase (EC 2.4.1.100), sucrose:fructan 6-fructosyltransferase (EC 2.4.1.10), fructan:fructan 6G-fructosyltransferase (EC 2.4.1.243) and levan fructosyltransferases (EC 2.4.1.-). The GH68 family consists of frucosyltransferases (FTFs) that include levansucrase (EC 2.4.1.10, also known as beta-D-fructofuranosyl transferase), beta-fructofuranosidase (EC 3.2.1.26) and inulosucrase (EC 2.4.1.9). GH32 and GH68 family enzymes are retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) and catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd09001 | GH43_FsAxh1-like | 0.002 | 327 | 423 | 140 | 224 | Glycosyl hydrolase family 43 such as Fibrobacter succinogenes subsp. succinogenes S85 arabinoxylan alpha-L-arabinofuranosidase. This glycosyl hydrolase family 43 (GH43) includes mostly enzymes that have been annotated as having beta-1,4-xylosidase (beta-D-xylosidase; xylan 1,4-beta-xylosidase; EC 3.2.1.37) activity. They are part of an array of hemicellulases that are involved in the final breakdown of plant cell-wall whereby they degrade xylan. They hydrolyze beta-1,4 glycosidic bonds between two xylose units in short xylooligosaccharides. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. This subfamily includes the characterized Clostridium stercorarium F-9 beta-xylosidase Xyl43B. It also includes Humicola insolens AXHd3 (HiAXHd3), a GH43 arabinofuranosidase (EC 3.2.1.55) that hydrolyzes O3-linked arabinose of doubly substituted xylans, a feature of the polysaccharide that is recalcitrant to degradation. It possesses an additional C-terminal beta-sandwich domain such that the interface between the domains comprises a xylan binding cleft that houses the active site pocket. The HiAXHd3 active site is tuned to hydrolyze arabinofuranosyl or xylosyl linkages, and the topology of the distal regions of the substrate binding surface confers specificity. It also includes Fibrobacter succinogenes subsp. succinogenes S85 arabinoxylan alpha-L-arabinofuranosidase (Axh1;Fisuc_1769;FSU_2269), Paenibacillus sp. E18 alpha-L-arabinofuranosidase (Abf43A), Bifidobacterium adolescentis ATCC 15703 double substituted xylan alpha-1,3-L-specific arabinofuranosidase d3 (AXHd3;AXH-d3;BaAXH-d3;BAD_0301;E-AFAM2), and Chrysosporium lucknowense C1 arabinoxylan hydrolase / double substituted xylan alpha-1,3-L-arabinofuranosidase (Abn7;AXHd). A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08999 | GH43_ABN-like | 0.007 | 334 | 405 | 155 | 222 | Glycosyl hydrolase family 43 protein such as endo-alpha-L-arabinanase. This glycosyl hydrolase family 43 (GH43) subgroup includes mostly enzymes with alpha-L-arabinofuranosidase (ABF; EC 3.2.1.55) and endo-alpha-L-arabinanase (ABN; EC 3.2.1.99) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. The GH43 ABN enzymes hydrolyze alpha-1,5-L-arabinofuranoside linkages while the ABF enzymes cleave arabinose side chains so that the combined actions of these two enzymes reduce arabinan to L-arabinose and/or arabinooligosaccharides. These arabinan-degrading enzymes are important in the food industry for efficient production of L-arabinose from agricultural waste; L-arabinose is often used as a bioactive sweetener. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV36630.1 | 0.0 | 1 | 494 | 1 | 494 |
| AIQ73487.1 | 0.0 | 1 | 494 | 1 | 494 |
| ASA21237.1 | 2.08e-298 | 1 | 489 | 1 | 487 |
| AJY73379.1 | 1.48e-279 | 1 | 488 | 1 | 484 |
| BCG59772.1 | 2.08e-277 | 1 | 489 | 1 | 486 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BYL_A | 3.24e-237 | 1 | 488 | 1 | 471 | ChainA, Levansucrase [Bacillus subtilis],3BYN_A Chain A, Levansucrase [Bacillus subtilis] |
| 3BYK_A | 6.52e-237 | 1 | 488 | 1 | 471 | ChainA, Levansucrase [Bacillus subtilis] |
| 3BYJ_A | 6.52e-237 | 1 | 488 | 1 | 471 | ChainA, Levansucrase [Bacillus subtilis] |
| 3OM7_A | 2.58e-232 | 29 | 488 | 1 | 453 | ChainA, Levansucrase [Priestia megaterium],3OM7_B Chain B, Levansucrase [Priestia megaterium],3OM7_C Chain C, Levansucrase [Priestia megaterium],3OM7_D Chain D, Levansucrase [Priestia megaterium] |
| 3OM6_A | 5.19e-232 | 29 | 488 | 1 | 453 | ChainA, Levansucrase [Priestia megaterium],3OM6_B Chain B, Levansucrase [Priestia megaterium],3OM6_C Chain C, Levansucrase [Priestia megaterium],3OM6_D Chain D, Levansucrase [Priestia megaterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P05655 | 2.17e-237 | 1 | 488 | 1 | 471 | Levansucrase OS=Bacillus subtilis (strain 168) OX=224308 GN=sacB PE=1 SV=1 |
| P21130 | 4.89e-236 | 1 | 488 | 1 | 471 | Levansucrase OS=Bacillus amyloliquefaciens OX=1390 GN=sacB PE=2 SV=1 |
| P94468 | 4.31e-230 | 1 | 488 | 1 | 471 | Inactive levansucrase OS=Geobacillus stearothermophilus OX=1422 GN=sacB PE=1 SV=1 |
| Q70XJ9 | 1.69e-83 | 64 | 484 | 278 | 700 | Levansucrase OS=Fructilactobacillus sanfranciscensis OX=1625 GN=levS PE=1 SV=1 |
| Q55242 | 3.09e-79 | 52 | 484 | 242 | 682 | Levansucrase OS=Streptococcus salivarius OX=1304 GN=ftf PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
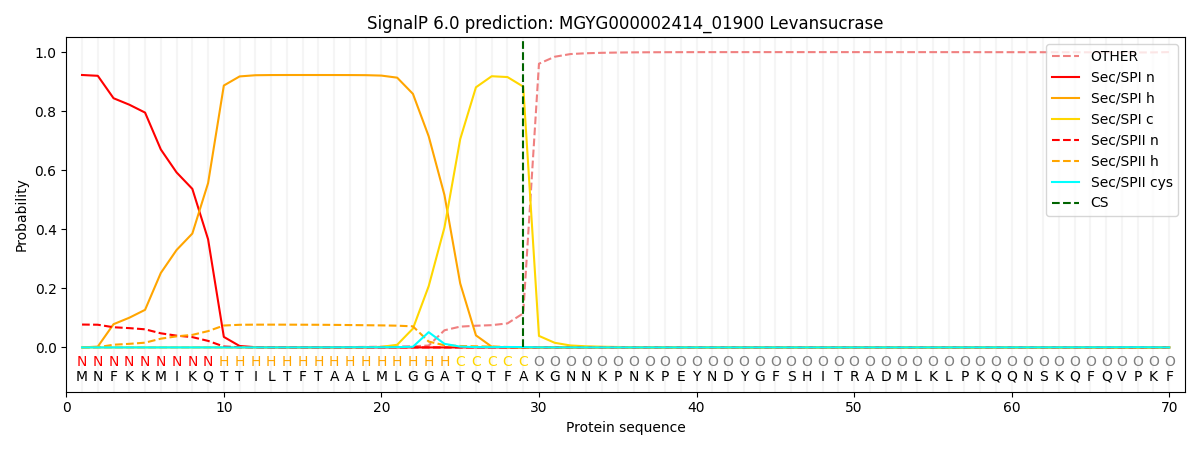
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000702 | 0.916841 | 0.081611 | 0.000328 | 0.000279 | 0.000219 |
