You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_02962
You are here: Home > Sequence: MGYG000002414_02962
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
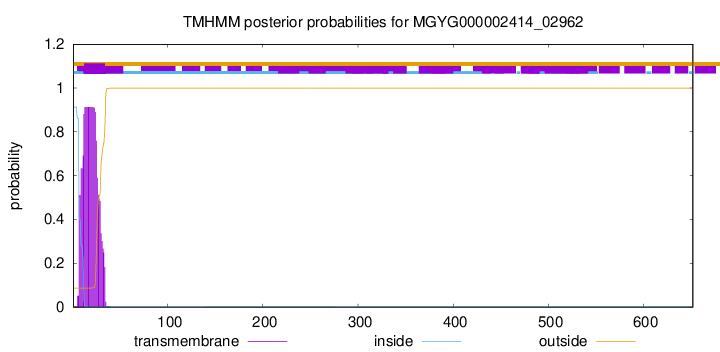
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_02962 | |||||||||||
| CAZy Family | GH93 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3343060; End: 3345018 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH93 | 50 | 339 | 6.9e-86 | 0.9674267100977199 |
| CBM13 | 363 | 507 | 1.5e-21 | 0.7021276595744681 |
| CBM13 | 513 | 649 | 1.1e-20 | 0.6861702127659575 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14200 | RicinB_lectin_2 | 1.37e-20 | 551 | 635 | 3 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 7.17e-20 | 451 | 542 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 2.23e-18 | 401 | 490 | 3 | 88 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.89e-16 | 497 | 590 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.45e-13 | 363 | 445 | 13 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV33771.1 | 0.0 | 1 | 610 | 1 | 610 |
| QGQ95143.1 | 1.96e-118 | 6 | 412 | 16 | 413 |
| AWF99909.1 | 2.19e-95 | 32 | 412 | 2 | 402 |
| QYX75859.1 | 4.82e-94 | 2 | 431 | 15 | 462 |
| CBG74713.1 | 2.30e-93 | 38 | 431 | 48 | 457 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2W5N_A | 4.97e-61 | 38 | 355 | 16 | 354 | ChainA, Alpha-l-arabinofuranosidase [Fusarium graminearum] |
| 2YDT_A | 2.61e-60 | 38 | 355 | 16 | 354 | ChainA, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],5M1Z_A Chain A, Exo-1,5-alpha-L-arabinofuranobiosidase [Fusarium graminearum] |
| 2W5O_A | 9.81e-60 | 38 | 355 | 16 | 354 | ChainA, ALPHA-L-ARABINOFURANOSIDASE [Fusarium graminearum] |
| 2YDP_A | 1.90e-59 | 38 | 355 | 16 | 354 | ChainA, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_B Chain B, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum],2YDP_C Chain C, Exo-1,5-alpha-l-arabinofuranobiosidase [Fusarium graminearum] |
| 3A71_A | 7.28e-58 | 38 | 353 | 11 | 341 | Highresolution structure of Penicillium chrysogenum alpha-L-arabinanase [Penicillium chrysogenum],3A72_A High resolution structure of Penicillium chrysogenum alpha-L-arabinanase complexed with arabinobiose [Penicillium chrysogenum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
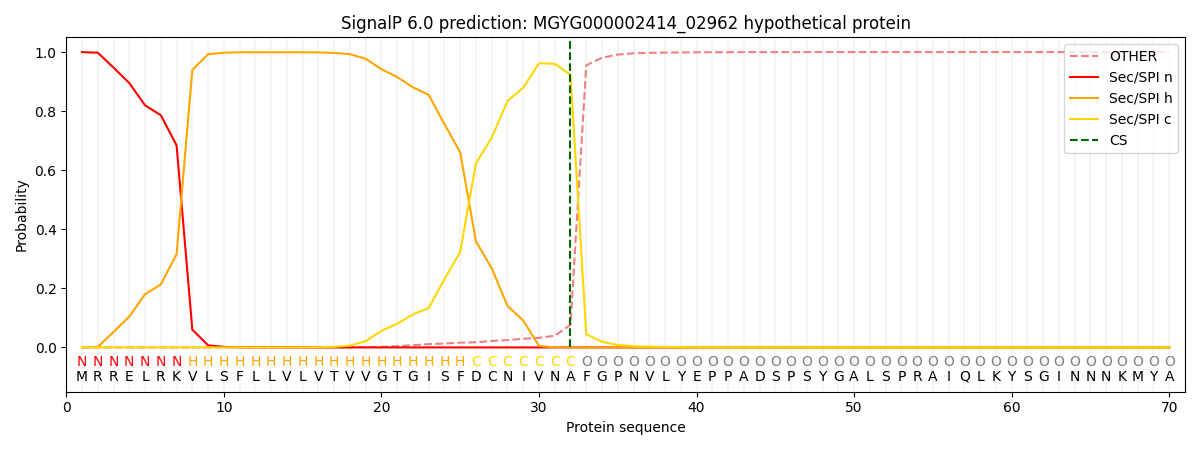
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000743 | 0.998364 | 0.000243 | 0.000253 | 0.000193 | 0.000170 |