You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_03056
You are here: Home > Sequence: MGYG000002414_03056
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_03056 | |||||||||||
| CAZy Family | CBM30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3458291; End: 3461641 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 318 | 768 | 6.6e-91 | 0.9952153110047847 |
| CBM30 | 66 | 235 | 3.2e-60 | 0.9941176470588236 |
| CBM3 | 977 | 1052 | 2.3e-26 | 0.9204545454545454 |
| CBM46 | 786 | 861 | 2.9e-16 | 0.9195402298850575 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 2.24e-72 | 322 | 767 | 3 | 374 | Glycosyl hydrolase family 9. |
| pfam03442 | CBM_X2 | 5.34e-33 | 779 | 861 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| pfam03442 | CBM_X2 | 2.16e-32 | 873 | 955 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| pfam00942 | CBM_3 | 5.06e-24 | 970 | 1051 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 7.85e-20 | 970 | 1052 | 1 | 83 | Cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV33854.1 | 0.0 | 1 | 1116 | 1 | 1116 |
| ANY67139.1 | 0.0 | 1 | 1115 | 1 | 1115 |
| QMV41506.1 | 0.0 | 167 | 1115 | 2 | 944 |
| ABN51859.1 | 4.66e-266 | 9 | 781 | 6 | 759 |
| ADU74665.1 | 4.66e-266 | 9 | 781 | 6 | 759 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3X17_A | 4.56e-75 | 217 | 768 | 16 | 554 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
| 1WMX_A | 4.37e-59 | 32 | 235 | 1 | 205 | ChainA, COG3291: FOG: PKD repeat [Acetivibrio thermocellus ATCC 27405],1WMX_B Chain B, COG3291: FOG: PKD repeat [Acetivibrio thermocellus ATCC 27405],1WZX_A Chain A, COG3291: FOG: PKD repeat [Acetivibrio thermocellus ATCC 27405],1WZX_B Chain B, COG3291: FOG: PKD repeat [Acetivibrio thermocellus ATCC 27405],1WZX_C Chain C, COG3291: FOG: PKD repeat [Acetivibrio thermocellus ATCC 27405],1WZX_D Chain D, COG3291: FOG: PKD repeat [Acetivibrio thermocellus ATCC 27405] |
| 5U0H_A | 8.00e-56 | 232 | 766 | 7 | 535 | Crystalstructure of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 5U2O_A | 8.80e-55 | 232 | 766 | 7 | 535 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 2C24_A | 9.56e-55 | 27 | 221 | 4 | 199 | ChainA, ENDOGLUCANASE [Acetivibrio thermocellus],2C24_B Chain B, ENDOGLUCANASE [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23659 | 1.86e-88 | 359 | 1115 | 65 | 986 | Endoglucanase Z OS=Thermoclostridium stercorarium OX=1510 GN=celZ PE=1 SV=1 |
| Q3MUH7 | 2.40e-73 | 868 | 1115 | 778 | 1027 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
| P50900 | 1.54e-68 | 860 | 1115 | 652 | 914 | Exoglucanase-2 OS=Thermoclostridium stercorarium (strain ATCC 35414 / DSM 8532 / NCIMB 11754) OX=1121335 GN=celY PE=1 SV=2 |
| Q02934 | 1.18e-52 | 368 | 1115 | 121 | 887 | Endoglucanase 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celI PE=1 SV=2 |
| Q05156 | 7.13e-52 | 219 | 770 | 185 | 742 | Cellulase 1 OS=Streptomyces reticuli OX=1926 GN=cel1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
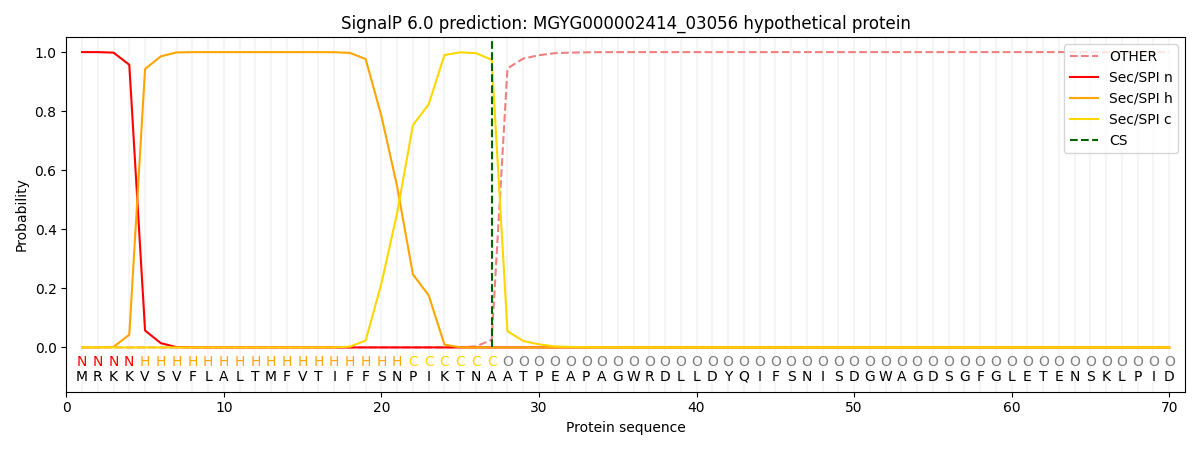
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000229 | 0.999052 | 0.000181 | 0.000183 | 0.000170 | 0.000151 |

