You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_03137
You are here: Home > Sequence: MGYG000002414_03137
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
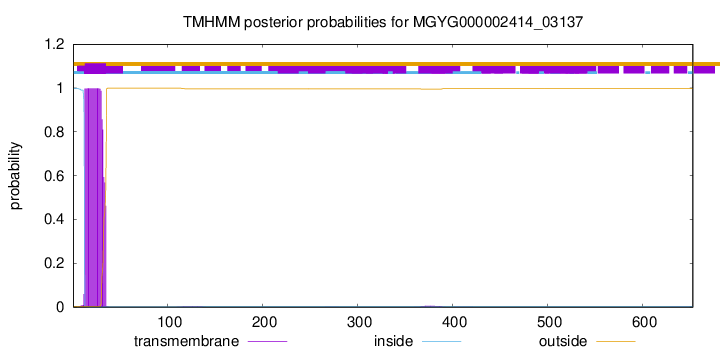
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_03137 | |||||||||||
| CAZy Family | GH8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3569273; End: 3571234 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.57e-20 | 532 | 645 | 1 | 123 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam01270 | Glyco_hydro_8 | 1.38e-16 | 112 | 374 | 33 | 277 | Glycosyl hydrolases family 8. |
| cd00057 | FA58C | 1.61e-14 | 535 | 652 | 16 | 143 | Substituted updates: Jan 31, 2002 |
| cd00063 | FN3 | 2.78e-12 | 433 | 517 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| COG3405 | BcsZ | 5.60e-12 | 68 | 321 | 19 | 254 | Endo-1,4-beta-D-glucanase Y [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV33925.1 | 0.0 | 1 | 653 | 1 | 653 |
| AIQ74597.1 | 0.0 | 1 | 653 | 1 | 653 |
| BAB64835.1 | 0.0 | 6 | 652 | 7 | 659 |
| AOO35455.1 | 5.18e-301 | 41 | 652 | 1 | 619 |
| QTH45936.1 | 1.51e-287 | 33 | 653 | 28 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CJU_A | 4.12e-208 | 36 | 423 | 2 | 390 | Crystalstructure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7CJU_B Crystal structure of inactive form of chitosanase crystallized by ammonium sulfate [Bacillus sp. K17-2],7XGQ_A Chain A, chitosanase [Bacillus sp. K17-2],7XGQ_B Chain B, chitosanase [Bacillus sp. K17-2] |
| 1V5C_A | 1.64e-207 | 41 | 423 | 1 | 384 | Thecrystal structure of the inactive form chitosanase from Bacillus sp. K17 at pH3.7 [Bacillus sp. (in: Bacteria)],1V5D_A The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)],1V5D_B The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 [Bacillus sp. (in: Bacteria)] |
| 5XD0_A | 2.45e-102 | 45 | 421 | 35 | 407 | ApoStructure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4],5XD0_B Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 [Paenibacillus sp. X4] |
| 2RV9_A | 8.27e-52 | 526 | 652 | 7 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 8.54e-52 | 526 | 652 | 8 | 137 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29019 | 4.12e-200 | 41 | 433 | 57 | 450 | Endoglucanase OS=Bacillus sp. (strain KSM-330) OX=72575 PE=1 SV=1 |
| P19254 | 2.66e-101 | 45 | 421 | 35 | 407 | Beta-glucanase OS=Niallia circulans OX=1397 GN=bgc PE=3 SV=1 |
| P37701 | 5.34e-35 | 41 | 420 | 32 | 389 | Endoglucanase 2 OS=Ruminiclostridium josui OX=1499 GN=celB PE=3 SV=1 |
| P37699 | 2.09e-33 | 38 | 420 | 29 | 389 | Endoglucanase C OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCC PE=1 SV=2 |
| A3DC29 | 1.32e-28 | 63 | 437 | 51 | 406 | Endoglucanase A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
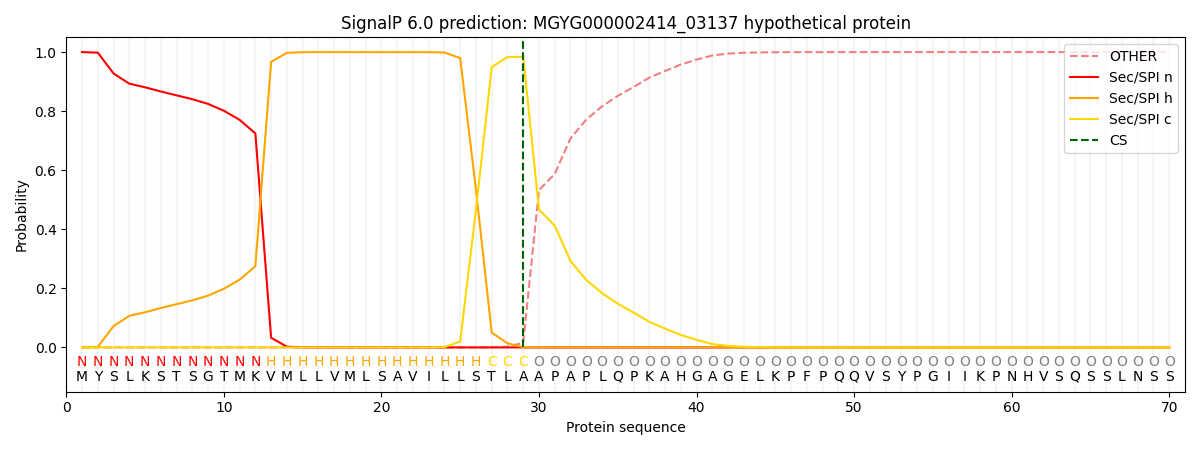
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.999209 | 0.000143 | 0.000145 | 0.000125 | 0.000120 |