You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_03480
You are here: Home > Sequence: MGYG000002414_03480
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
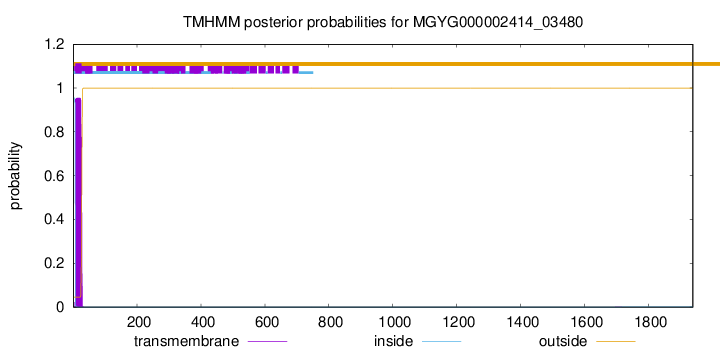
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_03480 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3938567; End: 3944386 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 1098 | 1468 | 2.7e-121 | 0.9733333333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07523 | Big_3 | 3.35e-11 | 619 | 685 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| NF033190 | inl_like_NEAT_1 | 1.83e-08 | 1759 | 1938 | 582 | 754 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam07523 | Big_3 | 1.03e-07 | 449 | 513 | 3 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam00395 | SLH | 8.49e-07 | 1759 | 1800 | 1 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 8.69e-06 | 1818 | 1859 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV34223.1 | 0.0 | 1 | 1939 | 1 | 1939 |
| ARK30471.1 | 0.0 | 2 | 1464 | 3 | 1476 |
| AOZ94532.1 | 0.0 | 211 | 1935 | 3 | 1692 |
| ADD02972.1 | 0.0 | 222 | 1936 | 18 | 1653 |
| QUW24060.1 | 0.0 | 153 | 1439 | 1 | 1272 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 1.32e-41 | 1103 | 1460 | 18 | 361 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 4.46e-13 | 1103 | 1446 | 6 | 390 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 6.16e-79 | 773 | 1385 | 33 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| C6CRV0 | 3.59e-43 | 1532 | 1935 | 1057 | 1458 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P0C1A7 | 1.25e-40 | 1103 | 1460 | 43 | 386 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 5.63e-40 | 1103 | 1460 | 43 | 386 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P38536 | 9.36e-29 | 1728 | 1936 | 1652 | 1855 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
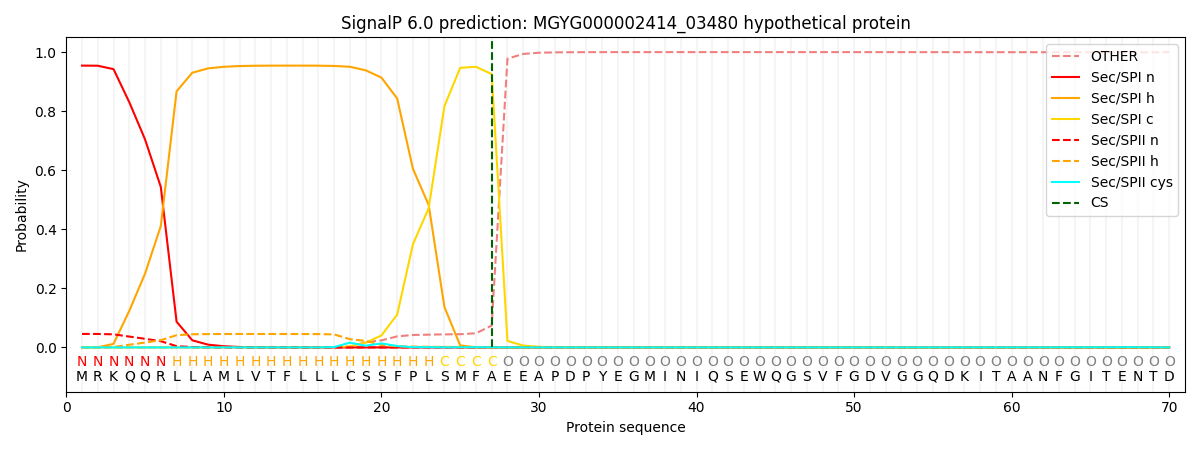
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000554 | 0.946850 | 0.051710 | 0.000296 | 0.000302 | 0.000263 |