You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_03795
You are here: Home > Sequence: MGYG000002414_03795
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
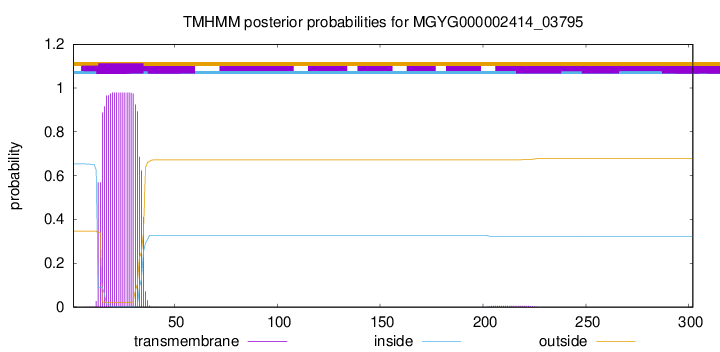
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_03795 | |||||||||||
| CAZy Family | GH46 | |||||||||||
| CAZyme Description | Chitosanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4292490; End: 4293398 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH46 | 63 | 287 | 3.3e-74 | 0.9954954954954955 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00978 | chitosanase_GH46 | 9.35e-61 | 66 | 289 | 1 | 221 | chitosanase belonging to the glycosyl hydrolase 46 family. This family is composed of the chitosanase enzymes which hydrolyzes chitosan, a biopolymer of beta (1,4)-linked-D-glucosamine (GlcN) residues produced by partial or full deacetylation of chitin. Chitosanases play a role in defense against pathogens such as fungi and are found in microorganisms, fungi, viruses, and plants. Microbial chitosanases can be divided into 3 subclasses based on the specificity of the cleavage positions for partial acetylated chitosan. Subclass I chitosanases such as N174 can split GlcN-GlcN and GlcNAc-GlcN linkages, whereas subclass II chitosanases such as Bacillus sp. no. 7-M can cleave only GlcN-GlcN linkages. Subclass III chitosanases such as MH-K1 chitosanase are the most versatile and can split both GlcN-GlcN and GlcN-GlcNAc linkages. |
| pfam01374 | Glyco_hydro_46 | 1.15e-21 | 75 | 256 | 2 | 178 | Glycosyl hydrolase family 46. This family are chitosanase enzymes. |
| cd16889 | chitinase-like | 7.51e-14 | 72 | 219 | 1 | 105 | chitinase-like domain. This family includes proteins such as chitinases, chitosanase, pesticin, and endolysin, which are involved in the degradation of 1,4-N-acetyl D-glucosamine linkages in chitin polymers and related activities. Chitinases are enzymes that catalyze the hydrolysis of the beta-1,4-N-acetyl-D-glucosamine linkages in chitin polymers. Chitosanase enzymes hydrolyze chitosan, a biopolymer of beta (1,4)-linked-D-glucosamine (GlcN) residues produced by partial or full deacetylation of chitin. Pesticin (Pst) is a anti-bacterial toxin produced by Yersinia pestis that acts through uptake by the target related bacteria and the hydrolysis of peptidoglycan in the periplasm. The dsDNA phages of eubacteria use endolysins or muralytic enzymes in conjunction with hollin, a small membrane protein, to degrade the peptidoglycan found in bacterial cell walls. Similarly, bacteria produce autolysins to facilitate the biosynthesis of its cell wall heteropolymer peptidoglycan and cell division. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV36800.1 | 3.67e-210 | 13 | 302 | 1 | 290 |
| AIQ75188.1 | 1.38e-205 | 13 | 302 | 1 | 290 |
| AIQ24850.1 | 1.13e-204 | 13 | 302 | 1 | 290 |
| QOS79836.1 | 2.73e-196 | 13 | 302 | 1 | 290 |
| ABC17783.1 | 1.29e-194 | 13 | 302 | 1 | 290 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1QGI_A | 3.26e-145 | 45 | 302 | 2 | 259 | ChainA, Protein (chitosanase) [Niallia circulans] |
| 2D05_A | 6.58e-145 | 45 | 302 | 2 | 259 | ChainA, Chitosanase [Niallia circulans] |
| 5HWA_A | 9.34e-145 | 45 | 302 | 2 | 259 | ChainA, Chitosanase [Niallia circulans] |
| 4ILY_A | 1.39e-08 | 62 | 226 | 16 | 169 | Abundantlysecreted chitosanase from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4ILY_B Abundantly secreted chitosanase from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 4OLT_A | 1.20e-06 | 68 | 257 | 21 | 205 | Chitosanasecomplex structure [Pseudomonas sp. LL2(2010)],4OLT_B Chitosanase complex structure [Pseudomonas sp. LL2(2010)] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33673 | 4.83e-145 | 42 | 302 | 41 | 301 | Chitosanase OS=Niallia circulans OX=1397 GN=csn PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
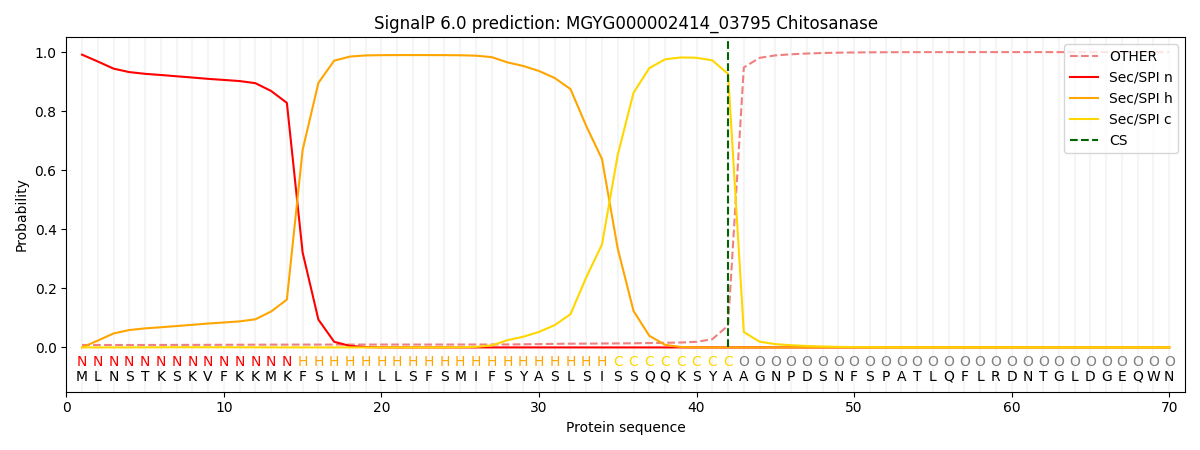
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009324 | 0.989387 | 0.000661 | 0.000261 | 0.000176 | 0.000169 |