You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_04001
You are here: Home > Sequence: MGYG000002414_04001
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_04001 | |||||||||||
| CAZy Family | CBM56 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4515872; End: 4517719 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM56 | 480 | 614 | 4.8e-37 | 0.949685534591195 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00063 | FN3 | 2.10e-14 | 432 | 516 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| COG3979 | COG3979 | 2.22e-13 | 431 | 523 | 4 | 97 | Chitodextrinase [Carbohydrate transport and metabolism]. |
| smart00060 | FN3 | 2.85e-12 | 432 | 503 | 1 | 81 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| pfam00041 | fn3 | 1.83e-09 | 434 | 504 | 2 | 81 | Fibronectin type III domain. |
| cd00146 | PKD | 0.001 | 432 | 514 | 1 | 81 | polycystic kidney disease I (PKD) domain; similar to other cell-surface modules, with an IG-like fold; domain probably functions as a ligand binding site in protein-protein or protein-carbohydrate interactions; a single instance of the repeat is presented here. The domain is also found in microbial collagenases and chitinases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV34692.1 | 0.0 | 1 | 615 | 1 | 615 |
| AIQ25044.1 | 0.0 | 1 | 615 | 6 | 620 |
| ANF94807.1 | 7.31e-293 | 4 | 614 | 3 | 532 |
| ATB31412.1 | 6.20e-256 | 33 | 614 | 49 | 630 |
| ATB44020.1 | 3.35e-228 | 35 | 614 | 52 | 629 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1K85_A | 9.09e-26 | 431 | 516 | 3 | 88 | ChainA, CHITINASE A1 [Niallia circulans] |
| 5T7A_A | 2.33e-23 | 523 | 613 | 25 | 116 | ChainA, BH0236 protein [Halalkalibacterium halodurans C-125],5T7A_B Chain B, BH0236 protein [Halalkalibacterium halodurans C-125] |
| 6BT9_A | 2.94e-11 | 363 | 506 | 429 | 556 | ChitinaseChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
| 6A6C_A | 2.82e-09 | 530 | 613 | 1 | 84 | Crystalstructure of carbohydrate-binding module family 56 beta-1,3-glucan binding domain [Paenibacillus barengoltzii] |
| 5H9X_A | 8.86e-08 | 525 | 613 | 18 | 107 | Crystalstructure of GH family 64 laminaripentaose-producing beta-1,3-glucanase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 9.62e-24 | 428 | 595 | 461 | 622 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P27050 | 1.47e-18 | 402 | 516 | 63 | 177 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| P50899 | 2.50e-15 | 423 | 548 | 695 | 815 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
| A0P8X0 | 3.52e-15 | 428 | 580 | 811 | 967 | Alpha-amylase OS=Niallia circulans OX=1397 GN=igtZ PE=1 SV=1 |
| P50401 | 1.42e-13 | 428 | 516 | 478 | 566 | Exoglucanase A OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
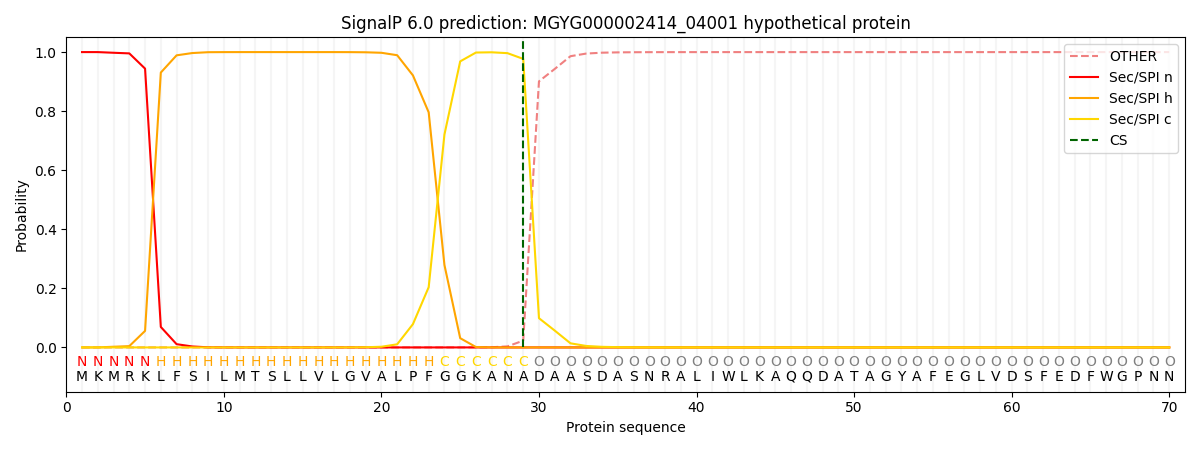
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000218 | 0.999150 | 0.000166 | 0.000177 | 0.000153 | 0.000136 |
