You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_04751
You are here: Home > Sequence: MGYG000002414_04751
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
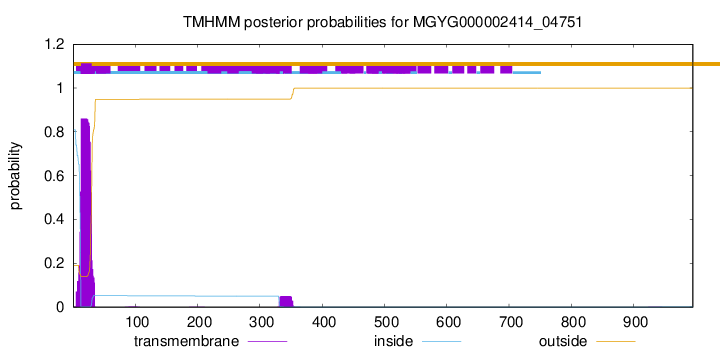
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_04751 | |||||||||||
| CAZy Family | GH48 | |||||||||||
| CAZyme Description | Exoglucanase-2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5318729; End: 5321716 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH48 | 43 | 500 | 8.3e-209 | 0.7299509001636661 |
| CBM3 | 851 | 931 | 1.5e-27 | 0.9772727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02011 | Glyco_hydro_48 | 0.0 | 44 | 739 | 4 | 619 | Glycosyl hydrolase family 48. Members of this family are endoglucanase EC:3.2.1.4 and exoglucanase EC:3.2.1.91 enzymes that cleave cellulose or related substrate. |
| pfam00942 | CBM_3 | 7.42e-26 | 849 | 930 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 5.83e-21 | 849 | 931 | 1 | 83 | Cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV36864.1 | 0.0 | 2 | 995 | 1 | 994 |
| AIQ76079.1 | 0.0 | 1 | 995 | 1 | 995 |
| AIQ54317.1 | 0.0 | 1 | 995 | 1 | 995 |
| ASA26471.1 | 0.0 | 2 | 995 | 1 | 994 |
| AIQ48863.1 | 0.0 | 2 | 995 | 1 | 994 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KW6_A | 0.0 | 42 | 743 | 4 | 704 | ChainA, Exoglucanase-2 [Bacillus licheniformis] |
| 5BV9_A | 0.0 | 40 | 741 | 2 | 701 | TheStructure of Bacillus pumilus GH48 in complex with cellobiose [Bacillus pumilus SAFR-032],5CVY_A The Structure of Bacillus pumilus GH48 in complex with cellobiose and cellohexaose [Bacillus pumilus SAFR-032],5VMA_A Structure of B. pumilus GH48 in complex with a cellobio-derived isofagomine [Bacillus pumilus] |
| 4EL8_A | 3.19e-211 | 44 | 742 | 12 | 633 | Theunliganded structure of C.bescii CelA GH48 module [Caldicellulosiruptor bescii DSM 6725] |
| 6D5D_A | 6.57e-211 | 44 | 742 | 23 | 644 | Structureof Caldicellulosiruptor danielii GH48 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 4L0G_A | 6.75e-211 | 44 | 742 | 16 | 637 | CrystalStructure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii DSM 6725],4L6X_A Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii DSM 6725],4TXT_A Crystal Structure of a GH48 cellobiohydrolase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P50900 | 1.55e-260 | 34 | 995 | 31 | 914 | Exoglucanase-2 OS=Thermoclostridium stercorarium (strain ATCC 35414 / DSM 8532 / NCIMB 11754) OX=1121335 GN=celY PE=1 SV=2 |
| P50899 | 3.45e-212 | 44 | 824 | 62 | 775 | Exoglucanase B OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhB PE=1 SV=1 |
| P22534 | 2.44e-197 | 44 | 742 | 1120 | 1741 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P37698 | 9.15e-197 | 8 | 736 | 6 | 652 | Endoglucanase F OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCF PE=1 SV=2 |
| P0C2S5 | 1.89e-195 | 9 | 732 | 5 | 653 | Cellulose 1,4-beta-cellobiosidase (reducing end) CelS OS=Acetivibrio thermocellus OX=1515 GN=celS PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
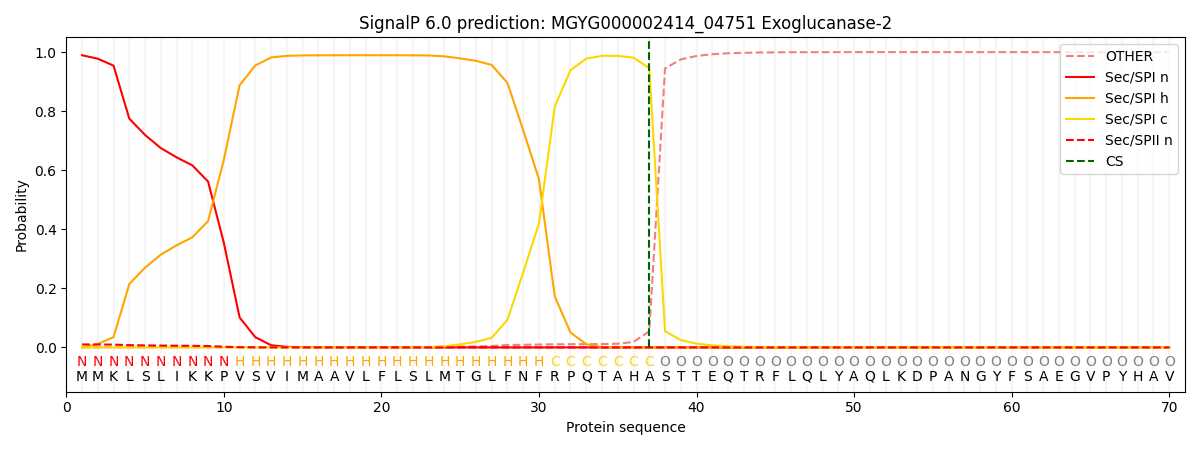
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001314 | 0.986390 | 0.011457 | 0.000368 | 0.000244 | 0.000196 |