You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002414_05612
You are here: Home > Sequence: MGYG000002414_05612
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus odorifer | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus odorifer | |||||||||||
| CAZyme ID | MGYG000002414_05612 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6296941; End: 6298377 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| SLH | 89 | 130 | 1.9e-19 | 0.9761904761904762 |
| SLH | 149 | 191 | 9.3e-16 | 0.9761904761904762 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00395 | SLH | 7.08e-13 | 89 | 130 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 2.05e-10 | 30 | 151 | 585 | 705 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 1.73e-06 | 32 | 71 | 2 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 4.18e-06 | 167 | 191 | 18 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 4.57e-04 | 74 | 190 | 567 | 682 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAU26536.1 | 6.06e-54 | 2 | 192 | 5 | 192 |
| QJD87768.1 | 5.65e-50 | 12 | 207 | 14 | 211 |
| BBH23221.1 | 4.65e-49 | 33 | 211 | 33 | 223 |
| BBH23452.1 | 1.84e-48 | 8 | 192 | 9 | 197 |
| ANY66681.1 | 3.93e-48 | 3 | 209 | 4 | 221 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 1.03e-10 | 30 | 195 | 29 | 192 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 1.04e-10 | 30 | 195 | 8 | 171 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 1.25e-21 | 33 | 195 | 1684 | 1851 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| Q06848 | 1.45e-20 | 51 | 206 | 242 | 405 | Cellulosome-anchoring protein OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=ancA PE=1 SV=1 |
| Q06852 | 1.66e-18 | 49 | 206 | 2101 | 2271 | Cell surface glycoprotein 1 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=olpB PE=3 SV=2 |
| C6CRV0 | 1.93e-17 | 38 | 195 | 1291 | 1455 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P19424 | 4.93e-16 | 38 | 197 | 49 | 212 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
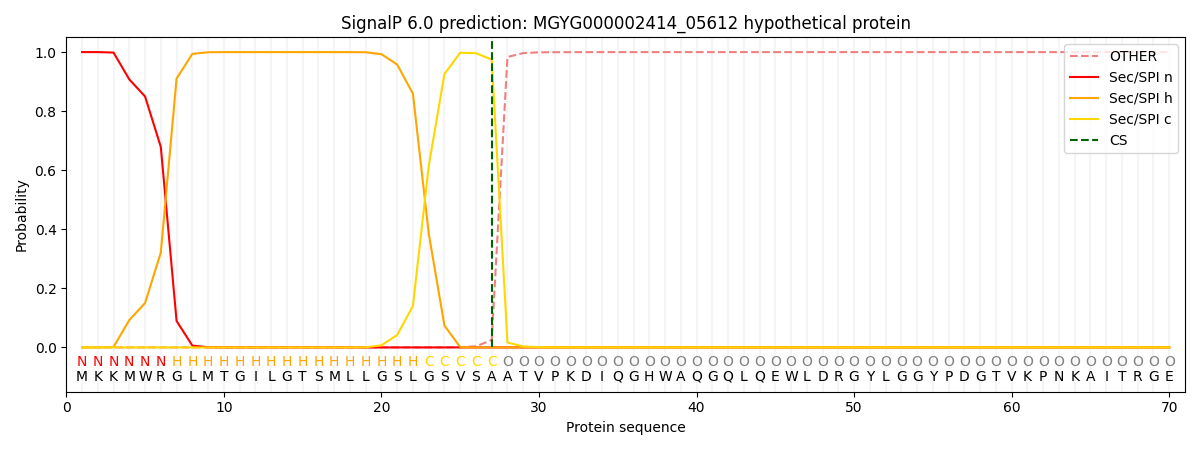
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999022 | 0.000191 | 0.000190 | 0.000179 | 0.000154 |
