You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002424_01244
You are here: Home > Sequence: MGYG000002424_01244
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
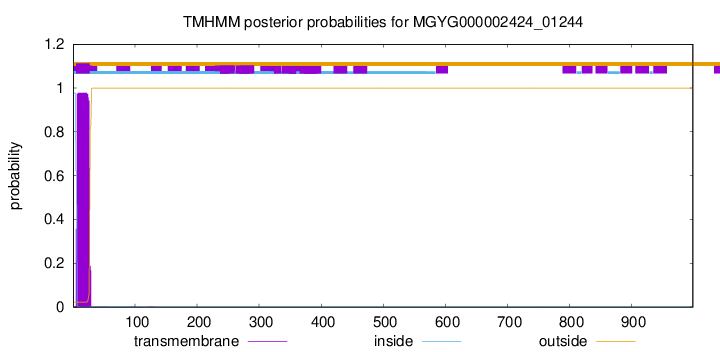
TMHMM annotations
Basic Information help
| Species | Monoglobus pectinilyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus pectinilyticus | |||||||||||
| CAZyme ID | MGYG000002424_01244 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1404077; End: 1407076 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 94 | 295 | 4e-68 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033761 | gliding_GltJ | 9.45e-09 | 813 | 917 | 395 | 489 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| NF033761 | gliding_GltJ | 2.22e-07 | 814 | 918 | 423 | 537 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| PHA03307 | PHA03307 | 1.75e-06 | 772 | 917 | 23 | 173 | transcriptional regulator ICP4; Provisional |
| NF033761 | gliding_GltJ | 2.03e-06 | 814 | 912 | 415 | 500 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| pfam03154 | Atrophin-1 | 2.43e-06 | 815 | 917 | 418 | 534 | Atrophin-1 family. Atrophin-1 is the protein product of the dentatorubral-pallidoluysian atrophy (DRPLA) gene. DRPLA OMIM:125370 is a progressive neurodegenerative disorder. It is caused by the expansion of a CAG repeat in the DRPLA gene on chromosome 12p. This results in an extended polyglutamine region in atrophin-1, that is thought to confer toxicity to the protein, possibly through altering its interactions with other proteins. The expansion of a CAG repeat is also the underlying defect in six other neurodegenerative disorders, including Huntington's disease. One interaction of expanded polyglutamine repeats that is thought to be pathogenic is that with the short glutamine repeat in the transcriptional coactivator CREB binding protein, CBP. This interaction draws CBP away from its usual nuclear location to the expanded polyglutamine repeat protein aggregates that are characteristic of the polyglutamine neurodegenerative disorders. This interferes with CBP-mediated transcription and causes cytotoxicity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19400.1 | 0.0 | 1 | 999 | 1 | 999 |
| AUO19402.1 | 2.34e-216 | 1 | 514 | 1 | 508 |
| AUO19476.1 | 1.34e-114 | 1 | 514 | 3 | 501 |
| AXP81340.1 | 1.30e-103 | 11 | 499 | 4 | 501 |
| AGA79324.1 | 1.07e-102 | 42 | 510 | 47 | 494 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 4.44e-52 | 37 | 500 | 17 | 410 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q2UB83 | 4.51e-50 | 37 | 500 | 17 | 410 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 2.09e-49 | 40 | 500 | 20 | 410 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| Q5B297 | 4.05e-49 | 40 | 500 | 20 | 407 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| A1DPF0 | 9.76e-47 | 40 | 500 | 21 | 411 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
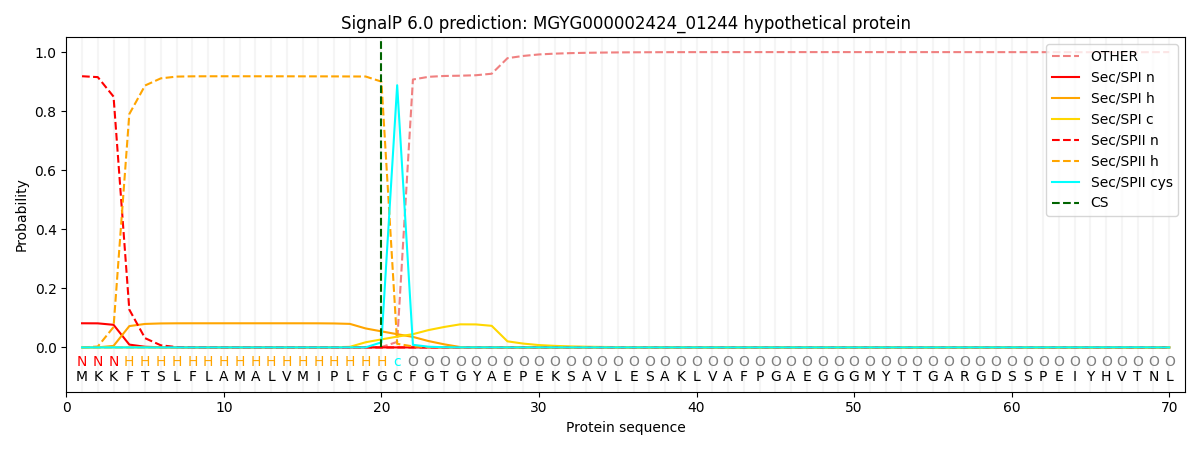
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000116 | 0.079801 | 0.919983 | 0.000039 | 0.000046 | 0.000038 |