You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002424_01448
You are here: Home > Sequence: MGYG000002424_01448
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Monoglobus pectinilyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus pectinilyticus | |||||||||||
| CAZyme ID | MGYG000002424_01448 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1678334; End: 1681936 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 93 | 293 | 3.4e-66 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18998 | Flg_new_2 | 3.29e-08 | 1008 | 1066 | 17 | 74 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| pfam04886 | PT | 9.52e-08 | 1067 | 1100 | 1 | 36 | PT repeat. This short repeat is composed on the tetrapeptide XPTX. This repeat is found in a variety of proteins, however it is not clear if these repeats are homologous to each other. The alignment represents nine copies of this repeat. |
| NF033186 | internalin_K | 9.64e-08 | 1000 | 1138 | 425 | 575 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
| COG3866 | PelB | 1.50e-07 | 38 | 279 | 27 | 272 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 3.55e-07 | 95 | 279 | 3 | 185 | Amb_all domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19604.1 | 0.0 | 1 | 1200 | 1 | 1200 |
| AUO18496.1 | 0.0 | 37 | 795 | 43 | 815 |
| AUO20150.1 | 0.0 | 6 | 795 | 7 | 796 |
| AUO20302.1 | 2.21e-266 | 16 | 807 | 6 | 854 |
| AUO20016.1 | 6.87e-227 | 25 | 799 | 19 | 791 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 2.57e-43 | 41 | 462 | 22 | 411 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 2.14e-42 | 21 | 462 | 5 | 411 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| Q2UB83 | 3.91e-42 | 41 | 462 | 22 | 411 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q5B297 | 1.04e-38 | 29 | 462 | 3 | 408 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| B0XMA2 | 8.38e-38 | 41 | 462 | 23 | 412 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
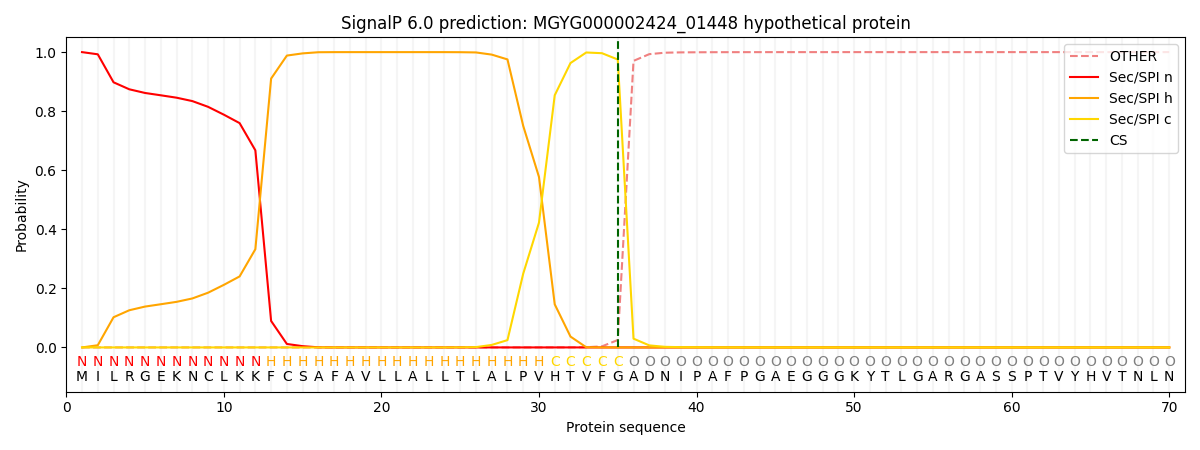
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000389 | 0.998903 | 0.000191 | 0.000173 | 0.000151 | 0.000144 |

