You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002424_01634
You are here: Home > Sequence: MGYG000002424_01634
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Monoglobus pectinilyticus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; Monoglobus; Monoglobus pectinilyticus | |||||||||||
| CAZyme ID | MGYG000002424_01634 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1939865; End: 1944154 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 1076 | 1386 | 1.4e-44 | 0.8541666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 1.58e-18 | 1171 | 1374 | 89 | 250 | pectinesterase |
| pfam02389 | Cornifin | 1.63e-18 | 593 | 715 | 13 | 134 | Cornifin (SPRR) family. SPRR genes (formerly SPR) encode a novel class of polypeptides (small proline rich proteins) that are strongly induced during differentiation of human epidermal keratinocytes in vitro and in vivo. The most characteristic feature of the SPRR gene family resides in the structure of the central segments of the encoded polypeptides that are built up from tandemly repeated units of either eight (SPRR1 and SPRR3) or nine (SPRR2) amino acids with the general consensus XKXPEPXX where X is any amino acid. In order to avoid bacterial contamination due to the high polar-nature of the HMM the threshold has been set very high. |
| PLN02682 | PLN02682 | 2.02e-17 | 1160 | 1374 | 140 | 315 | pectinesterase family protein |
| PLN02432 | PLN02432 | 6.13e-17 | 1225 | 1383 | 98 | 249 | putative pectinesterase |
| PLN02713 | PLN02713 | 2.55e-16 | 1177 | 1397 | 333 | 522 | Probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO19782.1 | 0.0 | 1 | 1429 | 1 | 1429 |
| AUO19003.1 | 5.99e-270 | 1 | 1425 | 1 | 1805 |
| AUO19783.1 | 1.34e-222 | 1 | 1370 | 1 | 1331 |
| AUO19460.1 | 2.84e-84 | 702 | 1386 | 1304 | 1934 |
| ADU21605.1 | 5.40e-67 | 1072 | 1429 | 366 | 721 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3UW0_A | 1.76e-14 | 1079 | 1376 | 43 | 309 | Pectinmethylesterase from Yersinia enterocolitica [Yersinia enterocolitica subsp. enterocolitica 8081] |
| 5C1E_A | 1.59e-13 | 1074 | 1401 | 13 | 272 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 5C1C_A | 2.14e-13 | 1235 | 1401 | 114 | 272 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
| 2NSP_A | 1.12e-11 | 1078 | 1376 | 16 | 285 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1GQ8_A | 1.66e-09 | 1164 | 1389 | 73 | 268 | Pectinmethylesterase from Carrot [Daucus carota] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SRX4 | 5.24e-17 | 1177 | 1397 | 346 | 535 | Probable pectinesterase/pectinesterase inhibitor 7 OS=Arabidopsis thaliana OX=3702 GN=PME7 PE=2 SV=1 |
| Q8RXK7 | 1.56e-16 | 1169 | 1397 | 328 | 529 | Probable pectinesterase/pectinesterase inhibitor 41 OS=Arabidopsis thaliana OX=3702 GN=PME41 PE=2 SV=2 |
| P38535 | 3.02e-14 | 28 | 202 | 905 | 1082 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| A2QK82 | 4.85e-14 | 1050 | 1401 | 18 | 300 | Probable pectinesterase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pmeA PE=3 SV=1 |
| Q8VYZ3 | 8.58e-14 | 1218 | 1374 | 182 | 329 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
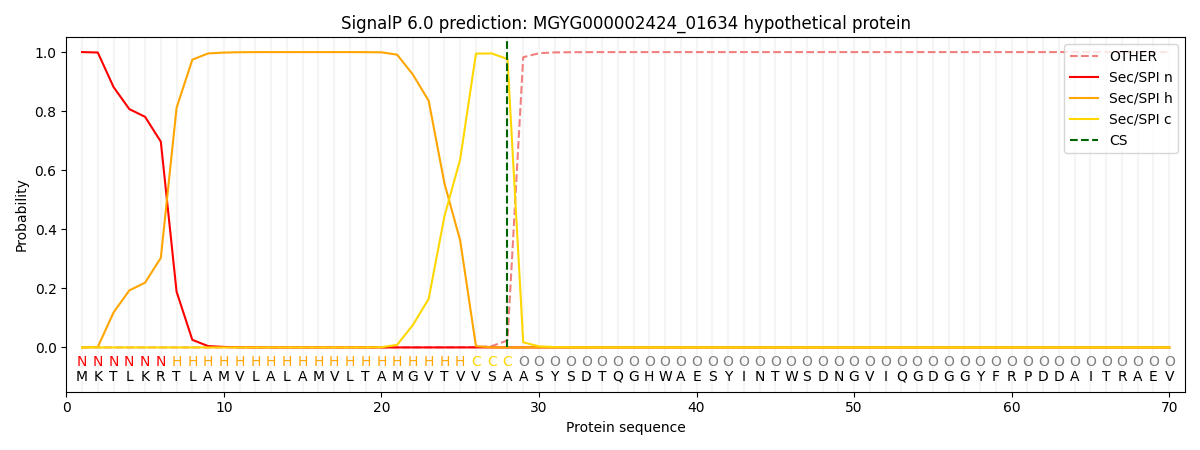
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000227 | 0.999122 | 0.000177 | 0.000173 | 0.000157 | 0.000137 |

