You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002438_00344
You are here: Home > Sequence: MGYG000002438_00344
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides distasonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides distasonis | |||||||||||
| CAZyme ID | MGYG000002438_00344 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 369976; End: 373314 Strand: + | |||||||||||
Full Sequence Download help
| MNSIKKMMLS GALGCLISGM AYADGSSPQP VKPYWQDIQV VAVNKEKPRS SFMSYADRET | 60 |
| ALTSRFEKSP YYSLLNGTWK FFFVDSYKDL PQNITDPSVN TSSWDDITVP GNWEVQGHGV | 120 |
| AIYTNHGYEF KPRNPQPPLL PEANPVGVYR RDIEIPASWD NRDIYLHIGG AKSGLYVYLN | 180 |
| GKEVGYSEDS KNPAEFLINK YLQPGKNVLT LKIFRWSTGS YLECQDFWRM SGIERDVFLW | 240 |
| SQPKASIQDF RVVSTLDDTY TNGIFKLAVD LKNHTQETKN LNVGYELLDA KGNLVTSEAN | 300 |
| DIWVSPASPQ TASFEYDLKN VAPWSAEHPN LYKLLMTIKE EGKVIEVVPF NVGFRRFEMK | 360 |
| QIDQVAEDGK PYTVLLFNGQ PVKFKGVNIH EHNPETGHYV TEELMRKDFE LMKQNNINAV | 420 |
| RLCHYPQDRK FYELCDEYGL YVYDEANIES HGMYYSLKKG GTLGNNPEWL IPHMDRTMNM | 480 |
| YERNKNYPSV TFWSLGNEAG NGYNFYQTYL YLKDKEINSM NRPVNYERAL WEWNTDMYVP | 540 |
| QYPSAEWLEE IGRKGSDRPV APSEYSHAMG NSSGNLWDQW KAIYKYPNLQ GGFIWDWVDQ | 600 |
| GILEKDKNGR EYYTYGGDYG VNAPSDGNFL CNGIVNPDRT PHPAMAEVKY AHQNIGFEAI | 660 |
| DLANGLFRVT NRFYFTNLKK YMVHYSVTAN NKVVRSGKVS LDIEPQASKE FTVPVGNLKP | 720 |
| QAGTEYFVNF NVTTVEKEPL IPIGHEIACD QFRLPIESPK KAFKTSGPKL TVSTNGDNLS | 780 |
| ASSSKVNFMF NKKTGIVTSY KVDGTEYFSE GFGIQPNFWR APTDNDYGNG MPKRLQVWKE | 840 |
| SSKNFNVTDA TVTMDGNNAV VNVNYLLPAG NLYIVNYTIY PSGAVNVAAR FTSTDMDAAQ | 900 |
| TEVSESTRTA TFTPGRDAAR KEASKLNVPR IGVRFRLPAS MNQVEYFGRG PAENYLDRNA | 960 |
| GSMVGLYKST AEELYFPYVR PQENGHHTDT RWVSLSTGKK GLLIQADNTI GFNALRNSIE | 1020 |
| DFDDEEATGL SRQWSNFTPE QVANHDEAAA KNVLRRQHHI NNITPRDFVE VCVDLKQQGV | 1080 |
| AGYDSWGSRP EPAYTLPANR EYNWGFTLIP LK | 1112 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 61 | 964 | 2.9e-209 | 0.9747340425531915 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09525 | lacZ | 0.0 | 35 | 1108 | 17 | 1024 | beta-galactosidase. |
| PRK10340 | ebgA | 0.0 | 35 | 1111 | 4 | 1000 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| COG3250 | LacZ | 2.34e-140 | 58 | 958 | 3 | 800 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| smart01038 | Bgal_small_N | 1.84e-104 | 782 | 1108 | 1 | 272 | Beta galactosidase small chain. This domain comprises the small chain of dimeric beta-galactosidases EC:3.2.1.23. This domain is also found in single chain beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 9.40e-90 | 374 | 657 | 7 | 302 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT19668.1 | 0.0 | 1 | 1112 | 1 | 1112 |
| QIX66118.1 | 0.0 | 1 | 1112 | 1 | 1112 |
| BBK89947.1 | 0.0 | 1 | 1112 | 3 | 1114 |
| QUT53259.1 | 0.0 | 1 | 1112 | 1 | 1112 |
| ABR42015.1 | 0.0 | 1 | 1112 | 1 | 1112 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 2.62e-235 | 33 | 1108 | 1 | 979 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 2.70e-235 | 33 | 1108 | 2 | 980 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 3DEC_A | 4.96e-214 | 32 | 1112 | 4 | 998 | ChainA, Beta-galactosidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3BGA_A | 2.49e-210 | 26 | 1112 | 2 | 1002 | Crystalstructure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],3BGA_B Crystal structure of beta-galactosidase from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 3MUY_1 | 1.10e-144 | 35 | 991 | 16 | 942 | Chain1, Beta-D-galactosidase [Escherichia coli K-12],3MUY_2 Chain 2, Beta-D-galactosidase [Escherichia coli K-12],3MUY_3 Chain 3, Beta-D-galactosidase [Escherichia coli K-12],3MUY_4 Chain 4, Beta-D-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 1.48e-234 | 33 | 1108 | 2 | 980 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| O52847 | 1.35e-220 | 26 | 1111 | 11 | 1034 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| A5F5U6 | 8.17e-171 | 35 | 1108 | 13 | 1016 | Beta-galactosidase OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=lacZ PE=3 SV=2 |
| Q9K9C6 | 9.18e-165 | 36 | 1108 | 11 | 1011 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| A1SWB8 | 3.31e-163 | 35 | 991 | 13 | 954 | Beta-galactosidase OS=Psychromonas ingrahamii (strain 37) OX=357804 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
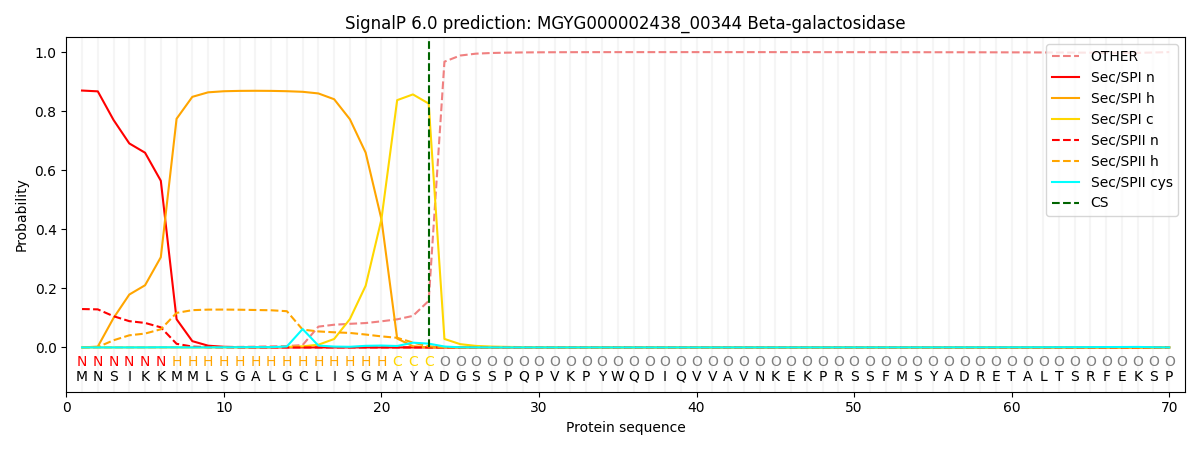
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001309 | 0.858556 | 0.139027 | 0.000467 | 0.000345 | 0.000278 |
