You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000002444_02199
You are here: Home > Sequence: MGYG000002444_02199
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Faecalibacillus faecis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Faecalibacillus; Faecalibacillus faecis | |||||||||||
| CAZyme ID | MGYG000002444_02199 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 41632; End: 45606 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 79 | 225 | 1.1e-26 | 0.9776119402985075 |
| CBM32 | 914 | 1028 | 3.9e-16 | 0.8548387096774194 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 8.36e-34 | 76 | 226 | 1 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.23e-19 | 79 | 226 | 6 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam00754 | F5_F8_type_C | 2.80e-09 | 924 | 1028 | 13 | 119 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00404 | Dockerin_1 | 3.76e-06 | 1267 | 1321 | 1 | 54 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| cd14256 | Dockerin_I | 4.82e-05 | 1267 | 1321 | 2 | 55 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL58565.1 | 0.0 | 1 | 1324 | 1 | 1328 |
| QTZ28728.1 | 3.26e-228 | 55 | 1131 | 113 | 1176 |
| SQG16429.1 | 3.26e-228 | 55 | 1091 | 115 | 1142 |
| QZA21247.1 | 4.22e-228 | 73 | 1091 | 135 | 1139 |
| SQF53247.1 | 4.22e-228 | 73 | 1091 | 135 | 1139 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JS4_A | 2.82e-11 | 78 | 226 | 612 | 752 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 2YGL_A | 1.88e-08 | 79 | 160 | 191 | 278 | TheX-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 [Streptococcus pneumoniae SP3-BS71],2YGL_B The X-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 [Streptococcus pneumoniae SP3-BS71],2YGM_A The X-Ray Crystal Structure Of Tandem Cbm51 Modules Of Sp3gh98, The Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae Sp3-Bs71, In Complex With The Blood Group B Antigen [Streptococcus pneumoniae SP3-BS71],2YGM_B The X-Ray Crystal Structure Of Tandem Cbm51 Modules Of Sp3gh98, The Family 98 Glycoside Hydrolase From Streptococcus Pneumoniae Sp3-Bs71, In Complex With The Blood Group B Antigen [Streptococcus pneumoniae SP3-BS71] |
| 2VMH_A | 2.69e-07 | 85 | 226 | 10 | 150 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMG_A | 3.03e-07 | 85 | 226 | 16 | 156 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2VMI_A | 4.96e-07 | 85 | 226 | 10 | 150 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
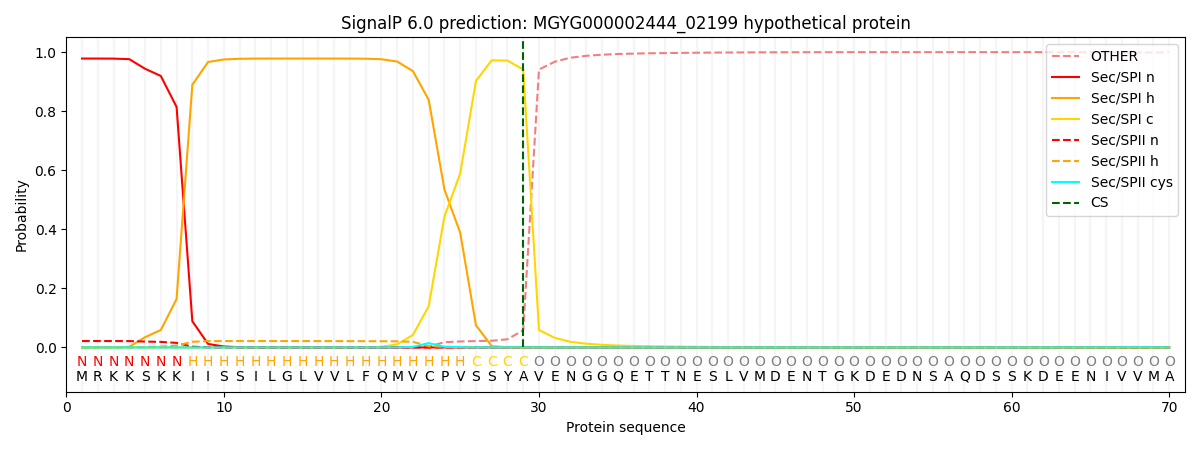
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001720 | 0.973640 | 0.023684 | 0.000345 | 0.000309 | 0.000275 |
